the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
On inference of boxplot symbolic data: applications in climatology
Abdolnasser Sadeghkhani
Ali Sadeghkhani
This paper presents a pioneering study on the inference of boxplot-valued data using both Bayesian and frequentist approaches within a multivariate framework. This approach leverages complex yet intuitive representations to make large datasets more manageable and enhance their interpretability, which is invaluable in the age of big data. Boxplot-valued data are particularly important due to their ability to capture the inherent variability and distributional characteristics of complex datasets.
In our study, we propose novel methodologies for parameter estimation and density estimation for boxplot-valued data and apply these techniques to climatological data. Specifically, we utilize data from the Berkeley Earth Surface Temperature Study, which aggregates 1.6 billion temperature reports from 16 pre-existing archives affiliated with the Lawrence Berkeley National Laboratory. Our methods are validated through extensive simulations comparing the efficiency and accuracy of Bayesian and frequentist estimators.
We demonstrate the practical applicability of our approach by analyzing summer average temperatures across various European countries. The proposed techniques provide robust tools for analyzing complex data structures, offering valuable insights into climatic trends and variations. Our study highlights the advantages and limitations of each inferential method, offering guidance for future research and applications in the field of climatology.
- Article
(833 KB) - Full-text XML
- BibTeX
- EndNote
Data analysts are now encountering increasingly large datasets whose analysis, spanning decades or even centuries, presents a complex challenge that demands more in-depth attention. There are many instances where complex and vast information, due to its intrinsic structure, cannot be adequately represented as single-valued data. As a result, symbolic data analysis (SDA) has emerged as a crucial approach, integrating elements of data science, multivariate analysis, pattern recognition, data mining, and artificial intelligence to analyze such data properly without losing information. Initially conceptualized by Diday (1988) and later formalized by Billard and Diday (2003), SDA represents a significant advancement in managing, reducing, and interpreting large datasets.
The initial steps of summarizing data to facilitate a clustering process applied to a large database appeared in the work of Diday and Noirhomme-Fraiture (2008), which, by introducing the cluster as a category, highlighted the importance of data summarization using SDA. To manage data effectively, SDA works without losing information, a common issue in traditional analyses, and encompasses various data formats, including intervals, sets, lists, histograms, trees, boxplots, and other distributional representations. By employing these complex yet intuitive representations, SDA not only makes vast amounts of data more manageable but also enhances their comprehensibility, proving to be an invaluable tool in the age of big data.
The importance of boxplot-valued data in real-world applications is particularly evident in climate science, where data often span long periods and involve significant uncertainty. For example, long-term datasets such as historical temperature records or remote sensing data generate vast volumes of observations that are often summarized as medians, quartiles, and extreme values for computational efficiency. These boxplot-valued summaries enable researchers to analyze trends in temperature variability, extreme events, or regional shifts in central tendencies, all of which are critical for understanding climate dynamics and making policy decisions.
By expanding data and converting their form from single-valued to symbolic (e.g., multivalued data such as list data, model-valued data such as histograms, or interval-valued data), the need for further theoretical development of SDA in each of its types has emerged. Diday (1995), Émilion (1997), and Diday (1988) provided the foundational mathematical background for various forms of symbolic data.
Among symbolic data types, interval-valued data have received significant attention due to their simplicity, versatility, and prevalence in various applied sciences compared to other symbolic data types. These data types, which range from univariate to multivariate, address the needs of real-world applications. In recent years, both frequentist and Bayesian approaches to interval-valued data estimators have been explored (e.g., Xu and Qin, 2022; Samadi et al., 2024; Sadeghkhani and Sadeghkhani, 2024). Additionally, various statistical tools for investigating relationships within interval-valued data have been developed. For instance, principal component analysis (PCA) using vertices to represent intervals was proposed by Douzal-Chouakria et al. (2011), a regression method that avoids the center and range approach was introduced by Billard and Diday (2000) and later by Neto and De Carvalho (2008) and Neto and De Carvalho (2010), and interval-valued time series methods were implemented by Xiong et al. (2015).
Beyond interval-valued data, boxplot-valued data provide additional distributional insights by including medians, quartiles, and extremes, making them well suited to summarizing massive datasets. In climate science, examples include datasets where raw observations (e.g., hourly or daily temperatures) are summarized into annual or monthly boxplots for computational feasibility. Historical datasets from pre-digital eras often come as summarized statistics (e.g., medians and extremes), making boxplot-valued data analysis indispensable. Such datasets allow for the study of trends in variability and centrality, which are critical for analyzing long-term climate change impacts.
The relevance of boxplot-valued data is also evident in studies of regional variability, where data collection is aggregated to minimize measurement noise. For example, climate model datasets often summarize temperature and precipitation data across spatial grids, which could naturally be represented and analyzed as boxplot-valued data. This approach reduces computational complexity while preserving the essential distributional features of the data.
Through this work, we aim to develop a methodological framework for analyzing boxplot-valued data and providing tools that are computationally efficient and applicable to a wide range of fields, including but not limited to climate science. By addressing the unique challenges posed by such data, we contribute to the broader goal of managing and interpreting massive datasets in the applied sciences.
While interval-valued data capture the range of variability, boxplot-valued data provide a richer summary by including additional distributional characteristics such as quartiles and the median. Boxplots are widely used for comparing distributions across different groups, detecting outliers, and understanding the spread and symmetry of the data. The concept of boxplots was first proposed by Tukey (1977) and further developed by Benjamini (1988). However, the mathematical foundation for statistical inference using boxplot-valued data has not been assessed thoroughly.
Though boxplots are valuable for comparing distributions across different groups, detecting outliers, and understanding the spread and symmetry of the data, their mathematical foundation has not been assessed thoroughly. They are widely used in various fields due to their simplicity and effectiveness in summarizing complex datasets. The concept of boxplots was first proposed by Tukey (1977) and was developed by Benjamini (1988). The importance of data visualization was further emphasized by Chambers (2018), who explored graphical methods for data analysis. Wickham and Wickham (2016) focused on modern approaches to creating boxplots using the ggplot2 package in R, highlighting practical aspects of their implementation.
To compare the role of boxplot-valued data, Arroyo et al. (2006) contrasted these types of data with interval-valued data and histograms. Boxplot variables serve as an intermediate point between the simplicity of interval variables and the detailed information provided by histogram variables. While interval variables do not convey information about the central area of an empirical distribution, boxplot variables do so using three quartiles. In contrast, histogram variables offer detailed insights into the empirical distribution, though their structure is more complex, requiring a set of consecutive intervals with associated weights. Despite their simpler structure, boxplot variables effectively capture the shape of the distribution.
To the best of our knowledge, there are no specific mathematical considerations for boxplot-valued data parameter estimation in the context of symbolic data, whether from a frequentist or Bayesian perspective. However, Reyes et al. (2024) proposed a parameterized regression method for boxplot-valued data by applying a Box–Cox transformation. Additionally, Reyes et al. (2022) focused on forecasting time series of these types of data.
Let Xi be a boxplot-valued observation summarized by its five-number summary statistics: the minimum (ai), first quartile (q1i), median (mi), third quartile (q3i), and maximum (bi). This structure effectively captures the distributional characteristics of the data for each observation. To facilitate more precise parameter estimation, we decompose Xi into two components, i.e., , which focuses on the central tendency and the shape of the distribution, and , which represents the range, thus capturing the overall spread of the data. By separating these components, we are able to model the central distribution and variability independently, thereby enhancing the accuracy of our parameter estimates.
While Le-Rademacher and Billard (2011) focused on univariate interval-valued data, modeling the midpoints in a frequentist framework, we extend their methodology to handle boxplot-valued data. They assumed that the midpoints of intervals are normally distributed and derived maximum likelihood estimators for the mean and variance. By assuming a uniform distribution within each interval, we leverage the properties outlined by Le-Rademacher and Billard to justify the normality of , where, for instance, q1i is the midpoint between ai and mi=q2i.
However, it is important to note that this extension from univariate intervals to multivariate quartiles relies on specific assumptions. These include the assumption that the quartiles are derived from sufficiently large datasets where the central limit theorem (CLT) ensures the approximation of normality. Additionally, we assume that the data within the intervals are uniformly distributed. While this assumption is commonly employed in interval-valued data analysis, deviations from uniformity or smaller datasets could impact the validity of the multivariate normal model. In such cases, a larger Qi dataset would be required to approximate normality using the CLT.
In our approach, we model Qi as following a trivariate normal distribution 𝒩3(μ,Σ), where is the mean vector and Σ is the covariance matrix. This extension allows us to capture more detailed distributional characteristics inherent in boxplot data, offering a balance between theoretical rigor and practical applicability.
Therefore, we have
and
where we assume that Qi (the quantiles) and Ri (the range) are conditionally independent under the assumption that the internal distribution (quantiles) of the interval is not directly driven by the overall width of the interval.
It is worth noting that, in our model, the range is assumed to follow a uniform distribution over the interval . This assumption is commonly employed in the context of interval-valued data analysis (e.g., Neto and De Carvalho, 2008; Zhao et al., 2023). The uniform distribution is chosen due to its simplicity and practical effectiveness, particularly when there is no prior information suggesting a different underlying distribution. The bounds Rmin and Rmax are determined empirically based on the observed data, ensuring that they capture the full range of variability in our dataset.
Note that, by defining the scaling parameter λ, the bounds ai and bi are hence related to the quantiles q1i and q3i as
where , which is known as the interquartile range.
The rest of the paper is organized as follows. In Sect. 2, we elaborate on the maximum likelihood (ML) method for estimating the unknown parameters of a p-variate boxplot-valued random variable where p≥1. This section also includes a simulation to assess this method as well as the asymptotic distribution of the ML estimators. Section 3 deals with Bayesian estimation of the parameters and evaluates the proposed methods. Section 4 investigates the methods of density estimation in boxplot-valued data, ranging from plugin types to posterior predictive density estimators. In Sect. 5, we illustrate the practical utility and effectiveness of the proposed techniques in analyzing and interpreting complex boxplot-valued environmental data. Finally, we conclude with a discussion in Sect. 6.
The likelihood function of the unknown parameters , and λ based on a random sample of size n, for , assuming that Eqs. (1) and (2) are conditionally independent despite the inherent relationship between Ri and Qi, is given by
It can easily be seen that the ML estimators of μ are the sample means of q1i, mi, and q3i, i.e.,
The ML estimator of Σ is the sample covariance matrix
while the ML estimators for Rmin and Rmax are
The scaling parameter λ is estimated by minimizing an objective function. That is,
Our assumption of conditional independence between Ri and Qi is made to simplify the estimation process, recognizing that Ri and Qi are functionally related via the quartiles and the scaling parameter λ. However, by assuming that λ is constant across observations, we reduce the direct influence of Qi on Ri, making this approximation reasonable in practice.
The primary reason we applied an objective function for minimizing λ was to directly assess how well the model captures the relationship between the observed ranges Ri and the estimated upper quantiles q3i. Furthermore, while maximum likelihood estimation (MLE) is a robust method for parameter estimation, it relies on certain assumptions about the distributional properties of the data. Minimizing the objective function allows for greater flexibility in modeling this relationship without imposing strict distributional assumptions. Future work could explore models that explicitly incorporate this dependency, potentially enhancing the accuracy of the parameter estimates.
In the next subsection, we generalize the univariate X into the p-variate with for p>1, and we focus on estimating the multivariate boxplot data.
2.1 Multivariate ML estimators
Consider a p-variate boxplot-valued random variable , where each for and . Moreover, we assume that
such that
where is the mean vector with and
is the covariance matrix, with Σk,l representing the covariance between Qk and Ql for . Moreover, the range is uniformly distributed over .
In a similar fashion to the univariate case with p=1, to estimate the parameters in a multivariate setting, we maximize the likelihood function
where represents the multivariate normal density function and is the density function of the uniform distribution for each component Rij, with Rmin and Rmax denoting the vectors of the minimum and maximum values for each component j of the range.
This results in the ML estimator of the parameters as presented in the following theorem.
Theorem 1.
Consider a p-variate boxplot-valued random variable , where and . Suppose that
with for and . The ML estimators of the mean vector , where , are given by
The ML estimator of the covariance matrix Σ is
The ML estimators of the range parameters are
and the ML estimator of the scaling parameter λj is
Proof. The proof involves straightforward maximization of the likelihood function. For brevity, it is omitted here.
□
The following subsection details the simulation setup for the proposed MLE method for symbolic boxplot-valued data.
2.2 Simulation study of ML estimators
To demonstrate the estimation procedure, we simulate data for p=3 variables with a sample size of n=100 and the following true parameters:
The true values for the scaling parameters are λ1=0.48, λ2=0.49, and λ3=0.51.
In the simulation, we generate the components Qi and Ri separately to simplify the estimation. Specifically, is generated from a trivariate normal distribution 𝒩3(μ,Σ) as outlined in Eq. (1), and is assumed to follow a uniform distribution over , as given in Eq. (2). This approach enables independent handling of each component's characteristics.
The mean squared error (MSE) is computed for the mean vector μ and the covariance matrices Σj,k for , as follows:
and
where and represent the (a,b)th elements of the estimated and true covariance matrices, respectively, in the ith simulation.
Table 1 presents the MLE parameters along with their MSEs. Note that the MSEs for λj () are reported as “NA”, indicating no variation in these parameters across simulations.
2.3 Asymptotic distribution of ML estimators for boxplot-valued data
In this subsection, we derive the asymptotic distribution of the ML estimators for the parameters of a boxplot-valued random variable.
Theorem 2.
Let denote the ML estimators for the parameters of boxplot-valued data. Under regularity conditions (see, e.g., Ferguson, 2017), the asymptotic distribution of the ML estimators is given as
where ℐ(θ) is the Fisher information matrix and is given by
with
where I3p is the 3p×3p identity matrix and ⊗ denotes the Kronecker product.
Proof. The Fisher information matrix is the expected value of the outer product of the score function , and it is given as
For the multivariate normal distribution , the score function for μ is
and hence
Similarly, for Σ, we have the score function
and therefore we have
Since , we can write
For the uniform distribution , the Fisher information for the parameter λj (related to the range) is derived as
This expression reflects the dependence of the Fisher information on the individual λj values.
Using the central limit theorem, the score function evaluated at the true parameter values θ is asymptotically normally distributed as
The covariance of the score function is related to the Fisher information matrix by the information matrix equality
The dimension of the parameter vector is given by
This completes the proof.
□
Sadeghkhani and Sadeghkhani (2024) studied the Bayesian inference of symbolic interval-valued data by introducing noninformative priors, including the Jeffreys prior, into the multivariate setting. In this section, we use informative priors for boxplot-valued data, which allows us to update our prior knowledge based on the observed data. We derive the posterior distributions and find the Bayes estimators under the squared error loss (SEL) criterion.
3.1 Likelihood and priors
Consider the model where are independent and identically distributed (iid) from 𝒩3p(μ,Σ) with additional parameters Rminj, Rmaxj, and λj for included in the likelihood function .
We assume the following priors for the parameters:
where μ0 is the prior mean, κ is the scaling factor, ν0 is the degrees of freedom, and Ψ0 is the scale matrix of the inverse Wishart distribution. Additionally, we have
where and dj are the bounds of the uniform distributions and αj and βj are the parameters of the Beta distribution for each j.
Next, we find the posterior distributions of the parameters in the following lemma.
Lemma 1 (posterior distributions). Under the given priors in Eqs. (19), (20), and (21) and the observed data , we have the following.
- i.
The posterior distribution of Σ is given by
where
and
- ii.
The posterior distribution of μ is
with
where represents a multivariate Student's t distribution with mean vector m, variance matrix A, and ν degrees of freedom.
- iii.
The posterior distribution of Rminj is given by
where Rminj,obs is the minimum observed Rminj.
- iv.
The posterior distribution of Rmaxj is given by
where Rmaxj,obs is the maximum observed Rmaxj.
- v.
The posterior distribution of λj is given by
Proof. According to Bayes' rule, the posterior distributions are derived as follows:
- i.
The posterior of Σ is given by
which is the kernel of an inverse Wishart distribution:
- ii.
The posterior of μ is given by
which results in a multivariate normal distribution for μ given Σ, i.e.,
By integrating out Σ, we have
- iii.
For Rminj, combining the likelihood with the uniform prior results in the posterior distribution
- iv.
For Rmaxj, combining the likelihood with the uniform prior results in the posterior distribution
- v.
For λj, combining the likelihood with the Beta prior results in the posterior distribution
□
Next, Theorem 3 provides the Bayes estimator of unknown parameters of a boxplot-valued random variable.
Theorem 3 (Bayes estimators under SEL). Under the assumptions of Lemma 1, the Bayes estimators under SEL, which are the means of the marginal posteriors, are obtained as follows:
- i.
The Bayes estimator of μ is
- ii.
The Bayes estimator of Σ is
where
and
- iii.
The Bayes estimators of Rminj and Rmaxj are
where Rminj,obs and Rmaxj,obs are the minimum observed Rminj and maximum observed Rmaxj, respectively.
- iv.
The Bayes estimator of λj is
Proof. The Bayes estimators are the means of the posterior distributions given in Lemma 1, and this completes the proof.
□
3.2 Simulation study in the Bayesian setup
Similar to Sect. 2.2, we conduct a simulation study to demonstrate the Bayesian estimation procedure. The simulation is carried out with the same setup and true values as described there, i.e., for p=3 variables with a sample size of n=100 and the true parameters specified in Eqs. (15)–(17).
Here, we use noninformative priors to ensure that the data predominantly influence the parameter estimates. In this simulation, a normal prior distribution with mean vector 0 and covariance matrix 102 Ip is used for μ. An inverse Wishart prior distribution is employed for Σ with a scale matrix S0=Ip and degrees of freedom . For each λj, a uniform 𝒰(0,1) () is used, which covers a broad range of values to avoid imposing any strong constraints.
The results of the Bayesian estimation, including the MSEs for each parameter, are summarized in Table 2.
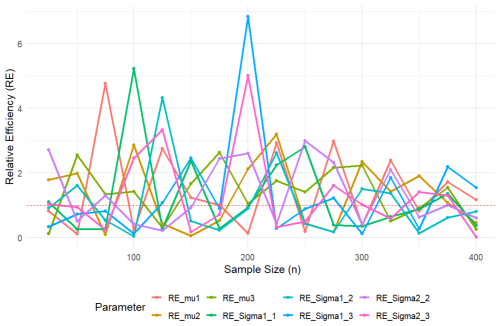
In order to compare the efficiency of the proposed ML and Bayesian estimators for different sample sizes n, we use the same simulation settings as those in Sects. 2.2 and 3.2. Figure 1 illustrates the relative efficiency (RE), defined as the MSE of the ML estimator over the MSE of the Bayesian estimator for the parameters μ and Σ. It can be seen that, for smaller sample sizes n, the efficiency of the Bayesian estimators is generally better (RE > 1). As n increases, thanks to the consistency of the ML estimators, the ML estimators tend to outperform the Bayesian estimators (RE < 1).
Given the posterior distributions of the parameters μ, Σ, λj, Rminj, and Rmaxj derived in Lemma 1, we can obtain the posterior predictive density for a new observation Q*. Theorem 4 finds the posterior predictive density estimator for a future or new random variable Q*.
Theorem 4 (posterior predictive density estimator). The posterior predictive density for a new random variable Q* given the observable is as follows:
- i.
When λj is known, the posterior predictive density estimator is given by
- ii.
When λj is unknown, the posterior predictive density is a mixture of multivariate Student's t distributions as given by
Proof. To derive the posterior predictive density, we need to integrate the parameters μ, Σ, and λj from the joint posterior distribution:
- i.
When λj is known, from Lemma 1, the posterior distributions of μ and Σ are given by
and therefore the posterior predictive density is given by
Since follows a normal inverse Wishart distribution, the resulting predictive density is a multivariate Student's t distribution:
- ii.
When λj is unknown, the posterior distributions of μ, Σ, and λj are
Then, the posterior predictive density is given by
and this completes the proof.
□
Remark 1. In addition to the posterior predictive density derived in Theorem 4, two other types of density estimators can be considered: plugin density estimators. We assume that λ is known or has been estimated using either Bayesian or ML methods.
- i.
ML plugin density estimator: by plugging in the ML estimators for μ, Σ, and λ from Theorem 2, the density estimator is given by
- ii.
Bayesian plugin density estimator: by plugging in the Bayesian estimators for μ, Σ, and λ from Theorem 3, the density estimator is
Note that, in both cases, λ influences the range but not the normal distribution part.
4.1 Comparison of density estimators using the expected Kullback–Leibler (KL) Loss
The KL loss function measures the difference between two probability distributions. For a true density p(Q) and an estimated density q(Q), the KL loss is defined as
The expected KL loss, also known as the KL risk function, for a density estimator q is the expectation of KL loss over the distribution of the data and is given as
To evaluate the performance of different predictive density estimators, we compare the KL risk performance of three methods in estimating the future density, i.e., Bayesian predictive, Bayesian plugin, and ML plugin estimators. The next lemma helps to find the KL risk functions.
Lemma 2. The KL loss between two multivariate normal distributions 𝒩p(μ1,Σ1) and 𝒩p(μ2,Σ2) is given by
The KL divergence between a multivariate normal distribution and a multivariate Student's t distribution is given by
where tr(⋅) denotes the trace of a given matrix.
Proof. The KL loss function between two multivariate normal densities is straightforward and therefore omitted. Given the density function of the multivariate normal distribution as
and the density function of the multivariate Student's t distribution as
substituting these into the KL loss function in Eq. (27) and simplifying them gives the result.
□
4.1.1 KL risk function comparison simulation
We conducted a simulation study to evaluate the KL risk for different sample sizes using three methods: Bayesian predictive, Bayesian plugin, and ML plugin estimators. The true parameters and hyperparameters used in the simulation are as follows. The true parameters are defined as
and the prior parameters are defined as
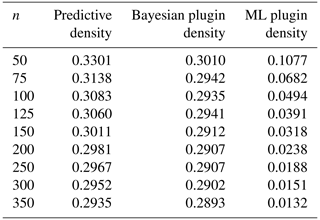
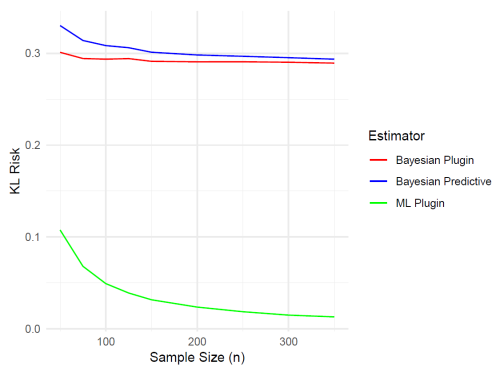
As seen from Table 3 and Fig. 2, the KL risks for the Bayesian predictive and plugin density estimators are quite close. Both risk functions are fairly small, and the difference between them decreases as the sample size n increases. Moreover, the KL risk for the ML plugin density estimator is smaller than that for the other two estimators, and it decreases as n increases. This suggests that the ML plugin density estimator performs better when estimating the new density. However, it is important to note that both Bayesian estimators were based on noninformative priors by choosing a very small value for κ0.
To visualize the comparison of KL divergence for different sample sizes, we present a plot in Fig. 2.
In this section, we apply the methodologies discussed – specifically the point estimation methods (both ML and Bayesian) and density estimations – to real climatological data. This approach demonstrates the practical utility and effectiveness of these statistical techniques in analyzing and interpreting complex boxplot-valued environmental data. The data are sourced from the Berkeley Earth Surface Temperature Study, which aggregates 1.6 billion temperature reports from 16 pre-existing archives affiliated with the Lawrence Berkeley National Laboratory (data can be found in the Berkeley Earth Surface Temperature Study at https://www.kaggle.com/berkeleyearth/climate-change-earth-surface-temperature-data, last access: 19 July 2024).
The section is divided into two subsections, each focusing on a distinct aspect of climatological analysis. First, we examine the monthly average temperatures in the United States from January 2000 to September 2013. In the latter subsection, we analyze summer average temperatures in European countries, with data going back to the 18th century. Both datasets are vast, containing extensive data. Aggregating these data into boxplots allows for computational efficiency by representing large amounts of information in a concise form.
5.1 Analysis of monthly average temperatures in the United States
5.1.1 Point estimators
The dataset used for this analysis includes monthly average temperatures from various cities across the United States, spanning from January 2000 to September 2013. These extensive data provide a comprehensive view of temperature trends and variations over a significant period. Here, each Xi represents the monthly average temperature summaries across multiple cities for month i within this period, capturing the variability of average temperatures in the form of symbolic data.
For each month, the first quartile (q1), median, and third quartile (q3) of the average temperatures across cities were calculated. Thus, each represents a vector of three summary statistics for month i, where the dimension of the data is defined as p=1, corresponding to the single temperature variable. These quartiles are then used to estimate the mean vector and covariance matrix using both ML and Bayesian methods.
The total number of monthly observations, n=165, spans from January 2000 to September 2013, resulting in 165 vectors of symbolic data, each representing monthly summary statistics of average temperatures. The hyperparameters for the Bayesian estimation are as follows: κ=0.001, , ν0=3, , a=0.3, b=0.4, c=1.8, and d=2.0.
Table 4 summarizes the results from both the ML and Bayesian estimations.
5.1.2 Density estimators
In this section, we explore three methods for estimating predictive densities: posterior predictive density, the ML method, and the Bayesian plugin method. These methods provide different approaches to predicting future monthly average temperatures in the United States.
The ML and Bayesian plugin density estimators, using the ML and Bayesian estimators for μ, Σ, and λ from Table 4, can be obtained easily and are denoted by and , respectively. On the other hand, the posterior predictive density for a new random variable Q* given the observations when λ is unknown is a mixture of multivariate Student's t distributions, as given in Eq. (26). If λ has already been estimated using ML or Bayesian methods, the posterior predictive density simplifies to a Student's t distribution as shown in Eq. (25).
5.2 Analysis of summer average temperatures in European countries
In this subsection, we extend our analysis to examine summer average temperatures across various European countries, utilizing historical climate data that provide comprehensive temperature records dating back to the 18th century. The dataset includes average monthly temperature data measured from numerous weather stations situated in major cities across Europe, allowing for an in-depth analysis of long-term climate trends and variations.
The analysis focuses on temperature records from 41 European countries, covering the three summer months June, July, and August. This extensive temporal and spatial coverage enables us to explore regional differences and similarities in temperature trends, contributing to a broader understanding of global climatic patterns.
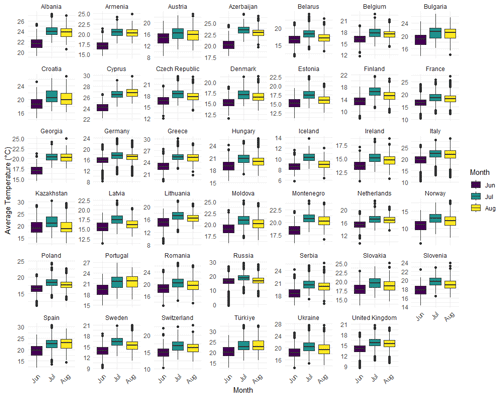
The dataset was filtered to include only European countries, with the average temperatures for the summer months extracted for further analysis. Boxplots were then generated to visualize the distribution of average temperatures across countries during the summer season. Each boxplot represents the distribution of average temperatures for a specific country across the three summer months.
Figure 3 presents these boxplots, providing a clear illustration of the temperature variations and trends across Europe. This comparative study highlights the regional climatic patterns, offering valuable insights into the broader context of global climate change.
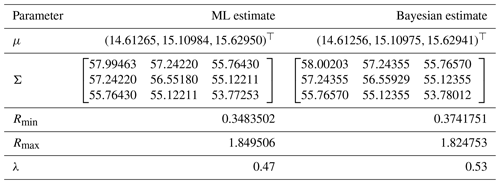
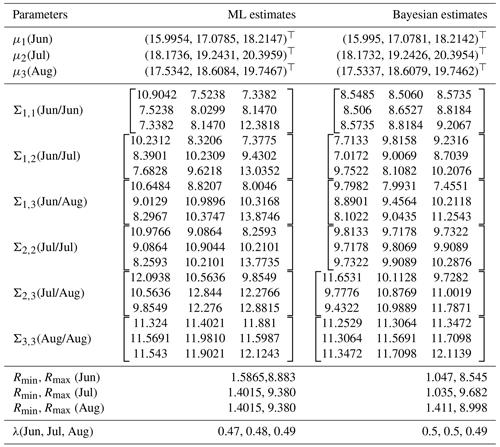
To further understand the differences between the ML and Bayesian approaches in estimating summer average temperatures in European countries, we present a comparative analysis of the estimators for the mean vectors and covariance matrices across the three summer months: June, July, and August. Table 5 summarizes the results.
The table above presents a side-by-side comparison of the ML and Bayesian estimators for the mean vectors and covariance matrices of summer average temperatures in European countries. The results show a close alignment between the ML and Bayesian estimates for the mean vectors across the three summer months. However, the covariance matrices exhibit some differences, particularly in the off-diagonal elements, which suggest variations in how each method captures the relationships between temperature readings across different months.
For instance, the Bayesian estimators tend to produce more consistent estimates across the covariance matrices, potentially reflecting the smoothing effect of prior information. The parameters Rmin and Rmax for each month also indicate that the range of temperature variations is slightly narrower in the Bayesian framework, further highlighting the influence of the priors.
In this article, we have introduced a novel method for estimating the parameters of boxplot-valued random variables from both Bayesian and frequentist perspectives. Working with boxplot-valued random variables enables us to efficiently summarize large datasets, as boxplots concisely represent extensive amounts of information. Our method offers a significant computational advantage, as we derived closed-form solutions for both ML and Bayesian estimators. These closed-form expressions allow for direct computation of parameter estimates without the need for iterative optimization procedures or Markov chain Monte Carlo (MCMC)-type methods, which are commonly required in traditional Bayesian approaches. By avoiding these computationally intensive steps, our method reduces the overall complexity and computational load, making it particularly suitable for large datasets.
Additionally, we have explored the density estimation of future random boxplots, providing a comprehensive tool for investigating and forecasting these distributions. Our approach was evaluated through simulations and demonstrated its accuracy and computational efficiency when compared with existing methods. Furthermore, we applied the proposed techniques to real environmental data to illustrate their practical utility and effectiveness in real-world scenarios.
Boxplot-valued data analysis is particularly useful in contexts where data are collected in large volumes and need to be summarized effectively without losing significant information. By adopting both Bayesian and frequentist methods, we provide a comprehensive framework that leverages the strengths of each approach. The Bayesian methods offer a probabilistic interpretation and incorporate prior information, which is valuable when data are scarce or expensive to obtain. On the other hand, the frequentist methods provide consistency and robustness when large amounts of data are available.
Furthermore, our study on the density estimation of future random boxplots enhances predictive modeling capabilities, which is crucial for applications such as environmental monitoring and climate forecasting. By accurately estimating the distribution of future data, decision-makers can make more informed predictions and plans.
Through extensive simulations, we validated the performance of our proposed methods, showcasing their accuracy and reliability. The application to real environmental data further underscores the practical relevance and adaptability of our techniques. Moreover, although the multivariate normal model theoretically allows for the possibility of overlapping quartiles, our empirical studies – including both simulations and real-world applications – consistently resulted in ordered quartiles (). This empirical consistency indicates that, within the context of our data and analysis, the model effectively preserves the natural ordering of quartiles, thereby mitigating the theoretical limitation in practical scenarios. Overall, this work contributes to the growing field of symbolic data analysis by offering efficient and effective tools for handling boxplot-valued data, thereby broadening the scope and applicability of statistical methodologies in various domains.
The R code used for the analysis in this study is not publicly available but can be shared upon reasonable request. Due to institutional policies and ongoing related research, we have opted not to deposit the code in a public repository at this time. However, researchers interested in accessing the code may contact the corresponding author at asadeghkhani@ncat.edu.
The data used in this study are publicly available and can be accessed from the Kaggle repository at https://www.kaggle.com/datasets/berkeleyearth/climate-change-earth-surface-temperature-data (Berkeley Earth, 2017). The dataset was obtained from this source and used in accordance with its terms of use. Researchers interested in the dataset can directly access it via the provided link.
AbS and AlS contributed equally to the development of the statistical methodology, study design, and data analysis. AbS prepared the initial manuscript draft, and both authors participated in its review and revision.
The contact author has declared that neither of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
The authors sincerely thank the editors and the two anonymous reviewers for their constructive feedback, which significantly improved the quality of this paper.
This paper was edited by Reetam Majumder and reviewed by two anonymous referees.
Arroyo, J., Maté, C., and Roque, A. M.-S.: Hierarchical clustering for boxplot variables, in: Data Science and Classification, edited by: Batagelj, V., Bock, H.-H., Ferligoj, A., and Žiberna, A., Springer, 59–66, https://doi.org/10.1007/3-540-34416-0_7, 2006. a
Benjamini, Y.: Opening the box of a boxplot, Am. Stat., 42, 257–262, 1988. a, b
Berkeley Earth: Climate Change: Earth Surface Temperature Data, Kaggle [data set], https://www.kaggle.com/datasets/berkeleyearth/climate-change-earth-surface-temperature-data (last access: 19 July 2024), 2017. a
Billard, L. and Diday, E.: Regression analysis for interval-valued data, in: Data analysis, classification, and related methods, edited by: Kiers, H. A. L., Rasson, J. P., Groenen, P. J. F., and Schader, M., Springer, 369–374, https://doi.org/10.1007/978-3-642-59789-3_58, 2000. a
Billard, L. and Diday, E.: From the statistics of data to the statistics of knowledge: symbolic data analysis, J. Am. Stat. Assoc., 98, 470–487, 2003. a
Chambers, J. M.: Graphical methods for data analysis, Chapman and Hall/CRC, https://doi.org/10.1201/9781351072304, 2018. a
Diday, E.: The symbolic approach in clustering and related methods of data analysis: the basic choices, in: Classification and Related Methods of Data Analysis, Proceedings of the First Conference of the International Federation of Classification Societies (IFCS-87: Technical University of Aachen, 673–684, North Holland, NII Article ID 10011477669, 1988. a, b
Diday, E.: Probabilist, possibilist and belief objects for knowledge analysis, Ann. Oper. Res., 55, 225–276, 1995. a
Diday, E. and Noirhomme-Fraiture, M.: Symbolic data analysis and the SODAS software, John Wiley & Sons, ISBN 978-0470018835, 2008. a
Douzal-Chouakria, A., Billard, L., and Diday, E.: Principal component analysis for interval-valued observations, Stat. Anal. Data Min., 4, 229–246, 2011. a
Émilion, R.: Différentiation des capacités et des intégrales de Choquet, C.R. Acad. Sci. I-Math., 324, 389–392, 1997. a
Ferguson, T. S.: A course in large sample theory, Routledge, ISBN 0412043718 2017. a
Le-Rademacher, J. and Billard, L.: Likelihood functions and some maximum likelihood estimators for symbolic data, J. Stat. Plan. Infer., 141, 1593–1602, 2011. a
Neto, E. d. A. L. and De Carvalho, F. D. A.: Centre and range method for fitting a linear regression model to symbolic interval data, Comput. Stat. Data An., 52, 1500–1515, 2008. a, b
Neto, E. d. A. L. and De Carvalho, F. D. A.: Constrained linear regression models for symbolic interval-valued variables, Comput. Stat. Data An., 54, 333–347, 2010. a
Reyes, D. M., de Souza, R. M., and de Oliveira, A. L.: A three-stage approach for modeling multiple time series applied to symbolic quartile data, Expert Syst. Appl., 187, 115884, https://doi.org/10.1016/j.eswa.2021.115884, 2022. a
Reyes, D. M., Souza, L. C., de Souza, R. M., and de Oliveira, A. L.: Parametrized linear regression for boxplot-multivalued data applied to the Brazilian Electric Sector, Inform. Sci., 652, 119758, https://doi.org/10.1016/j.ins.2023.119758, 2024. a
Sadeghkhani, A. and Sadeghkhani, A.: Multivariate Interval-Valued Models in Frequentist and Bayesian Schemes, arXiv [preprint], https://doi.org/10.48550/arXiv.2405.06635, 2024. a, b
Samadi, S. Y., Billard, L., Guo, J. H., and Xu, W.: MLE for the parameters of bivariate interval-valued model, Adv. Data Anal. Classif., 18, 827–850, https://doi.org/10.1007/s11634-023-00546-6, 2024. a
Tukey, J.: Exploratory data analysis, Springer, https://doi.org/10.1007/978-3-031-20719-8_2, 1977. a, b
Wickham, H. and Wickham, H.: Getting Started with ggplot2, ggplot2: Elegant graphics for data analysis, Springer, 11–31, https://doi.org/10.1007/978-3-319-24277-4_2, 2016. a
Xiong, T., Li, C., Bao, Y., Hu, Z., and Zhang, L.: A combination method for interval forecasting of agricultural commodity futures prices, Knowl.-Based Syst., 77, 92–102, 2015. a
Xu, M. and Qin, Z.: A bivariate Bayesian method for interval-valued regression models, Knowl.-Based Syst., 235, 107396, https://doi.org/10.1016/j.knosys.2021.107396, 2022. a
Zhao, Q., Wang, H., and Wang, S.: Robust regression for interval-valued data based on midpoints and log-ranges, Adv. Data Anal. Classi., 17, 583–621, 2023. a
- Abstract
- Introduction
- Maximum likelihood estimators
- Bayesian estimation for boxplot-valued data
- Posterior predictive density estimator
- Applications in climatology
- Discussion
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Review statement
- References
- Abstract
- Introduction
- Maximum likelihood estimators
- Bayesian estimation for boxplot-valued data
- Posterior predictive density estimator
- Applications in climatology
- Discussion
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Review statement
- References