the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Comparison of climate time series – Part 5: Multivariate annual cycles
Michael K. Tippett
This paper develops a method for determining whether two vector time series originate from a common stochastic process. The stochastic process considered incorporates both serial correlations and multivariate annual cycles. Specifically, the process is modeled as a vector autoregressive model with periodic forcing, referred to as a VARX model (where X stands for exogenous variables). The hypothesis that two VARX models share the same parameters is tested using the likelihood ratio method. The resulting test can be further decomposed into a series of tests to assess whether disparities in the VARX models stem from differences in noise parameters, autoregressive parameters, or annual cycle parameters. A comprehensive procedure for compressing discrepancies between VARX models into a minimal number of components is developed based on discriminant analysis. Using this method, the realism of climate model simulations of monthly mean North Atlantic sea surface temperatures is assessed. As expected, different simulations from the same climate model cannot be distinguished stochastically. Similarly, observations from different periods cannot be distinguished. However, every climate model differs stochastically from observations. Furthermore, each climate model differs stochastically from every other model, except when they originate from the same center. In essence, each climate model possesses a distinct fingerprint that sets it apart stochastically from both observations and models developed by other research centers. The primary factor contributing to these differences is the difference in annual cycles. The difference in annual cycles is often dominated by a single component, which can be extracted and illustrated using discriminant analysis.
Two fundamental questions arise repeatedly in climate science. (1) Has climate variability changed over time? (2) Do climate models accurately reflect reality? Answering these questions requires an objective procedure for deciding whether two time series possess identical statistical properties. Unfortunately, many procedures for deciding this have crucial limitations. Specifically, many of these procedures lack a significance test, do not account for serial correlation, or do not generalize naturally to multivariate quantities. For instance, the recent report from the Intergovernmental Panel on Climate Change frequently employed a quantity called RMSD (relative space–time root mean square deviation, Eyring et al., 2021, Sect. 3.8.2) to quantify the difference between simulated and observed seasonal cycles. However, RMSD lacks a rigorous significance test. Without an assessment of significance, one does not know whether a particular value of RMSD indicates genuine model deficiencies or is merely a result of random variations. One might attempt to adapt the F test to test significance, but many climate time series exhibit serial correlations, whereas the F test and other standard procedures assume that the data come from white noise processes. Applying a procedure that assumes white noise for a time series that is serially correlated leads to biased type-I errors. Furthermore, RMSD is evaluated for each physical variable separately. Often, RMSDs for different models and variables are displayed together in a checkerboard format (e.g., Fig. 3.42 of Eyring et al., 2021). A criterion for selecting a single winner when no single model produces the best RMSD for all the variables is rarely given.
The above limitations of RMSDs are also present in correlation skill and probabilistic verification measures. Although recent advancements in machine learning, as highlighted in works by Labe and Barnes (2022) and Brunner and Sippel (2023), have shown impressive capabilities for differentiating observations and simulations, these techniques also have no corresponding significance tests. While rigorous tests for comparing serially correlated, multivariate time series do exist (see Lund et al., 2009, for a lucid review), these too have shortcomings when applied specifically to climate time series. For instance, spectral-domain tests have less statistical power than time-domain tests (for further discussion, see DelSole and Tippett, 2020).
We have pursued an approach to comparing time series that avoids the above limitations. Specifically, we assume that each time series is generated by an autoregressive model. Under this assumption, two time series are said to originate from the same process if they share identical autoregressive model parameters. This paper represents the fifth installment in a series that develops this methodology. Part 1 laid the foundation for our approach to comparing univariate time series. Part 2 generalized this framework to multivariate processes, incorporating bias corrections to obtain statistics with chi-squared distributions. Part 3 developed methods to diagnose dissimilarities between two time series. This included a hierarchical testing procedure to attribute differences to specific components of the autoregressive model and a discriminant analysis technique to derive optimal diagnostics that contain all relevant information about differences between data sets. Part 4 incorporated annual cycles in the framework, which required generalizing the hierarchical testing procedure to include nonstationary forcing. However, this extension was limited to univariate time series.
In this paper, we generalize our test to account for multivariate periodic signals in the time series. To do this, we add periodic forcing to a vector autoregressive model to construct a VARX model, where X stands for exogenous variables such as periodic forcing. Then, the hypothesis that two VARX models share the same parameters is tested using the likelihood ratio method. If differences are detected, a stepwise procedure is employed to assess whether disparities in the VARX models stem from differences in noise parameters, autoregressive parameters, or periodic forcing parameters. To implement the test, we introduce three practical advances. First, we adjust maximum likelihood quantities to eliminate the biases mentioned earlier, which are particularly serious in multivariate problems. Second, we develop a Monte Carlo technique to determine significance thresholds. Third, diagnostics that maximally compress differences in VARX models into the fewest number of components are developed. Some of these diagnostics were developed in DelSole and Tippett (2022a). The extension of these diagnostics to arbitrary stepwise test procedures is completed in this paper. The present paper incorporates all of our “lessons learned” from previous parts into a single framework. Although this paper builds on our previous papers (DelSole and Tippett, 2020, 2021, 2022a, b), the present paper is nearly self-contained and can be understood largely on its own. Moreover, the code developed for this part supersedes our previous codes, as the previous codes are mere special cases of the general code provided with this paper.
Our problem is to decide whether two multivariate time series originated from the same stochastic process. Let the two time series be denoted as
where yt and are S-dimensional vectors, denoted as yt∈ℝS and . Here, S is the spatial dimension and N′ and denote the number of time samples. The random matrices Y and Y* are independent, but elements within each matrix may be correlated. That is, each time series may be spatially and serially correlated. We want to test the hypothesis that Y and Y* were generated independently by the same stochastic process.
A class of stochastic models that can capture multivariate serial correlations is a vector autoregressive (VAR) model. VAR models include linear inverse models (LIMs) as a special case, where LIMs are used extensively in seasonal and decadal prediction studies (Penland and Sardeshmukh, 1995; Whitaker and Sardeshmukh, 1998; Alexander et al., 2008; Vimont, 2012; Newman, 2013; Zanna, 2012). More general VAR models have also been used to assess climate predictability and causality (Mosedale et al., 2006; Chapman et al., 2015; Bach et al., 2019). Asymptotically, the statistics of VAR models are stationary (i.e., independent of t). Stationarity is a reasonable assumption for certain climate time series such as annual means (as analyzed in Parts 1–3). On the other hand, stationarity is violated for sub-annual time series that contain seasonal cycles or for multidecadal time series that contain climate trends.
To capture variations in the mean, such as annual cycles, we add deterministic forcing to the VAR model. The resulting model is called a VAR model with exogenous variables and is denoted as VARX (Lütkepohl, 2005, p. 387). This VARX model does not capture variations in covariances, although it can be generalized to do so (Lütkepohl, 2005, chapter 17). We will not pursue such generalizations here.
Mathematically, we assume each vector is generated by a VARX model of order p, with a deterministic forcing term ft∈ℝJ:
where the following matrices and vectors are constant for ,
and ϵt and are independent S-dimensional Gaussian white noise processes with respective covariance matrices Γ and Γ*. Because ϵt and are independent, so are yt and (although the latter vectors typically are separately serially correlated). The matrices are assumed to yield a stable VAR process, and similarly the matrices are assumed to yield a stable VAR process (the precise condition for stability is Eq. 2.1.12 in Lütkepohl, 2005). The matrices Γ and Γ* will be identified as noise parameters, and the matrices and will be identified as AR parameters.
In this paper, the deterministic term ft is associated with the annual cycle, and therefore C may be called the annual cycle parameters. These terms are simply sines and cosines with a period of 12 months and the associated harmonics (see Appendix A for an explicit definition). For such periodic forcing, long-term solutions of Eqs. (1) and (2) have the form of a time-periodic mean component accompanied by stationary noise centered around the mean. Notably, the nonstationarity is present solely in the means, while other long-term aspects of the system are stationary. The AR parameters describe time dependencies and are associated with the predictability or dynamics of the system. Thus, differences in AR parameters imply differences in dynamics or in predictability. On the other hand, differences in annual cycle parameters imply differences in annual cycle forcing, which should be distinguished from the annual cycle response, which depends on both the annual cycle and AR parameters. The annual cycle response for stable VAR parameters is derived by finding a solution that is exactly periodic on annual timescales and that can be expressed in closed form (see DelSole and Tippett, 2022b). Finally, noise parameters characterize variability after serial correlations and nonstationary signals have been removed. We call such variability whitened variability (also called one-step prediction errors), and hence differences in noise parameters imply differences in whitened variability.
To decide whether two multivariate time series came from the same stochastic process, we test the null hypothesis that the parameters of the two VARX(p) models are equal. More precisely, we test the hypothesis
The intercept terms d and d* are not included in H0. The reason for excluding the intercept term in our particular comparison is that this term controls the climatological mean, and climate models are known to have large biases. There is little value in assessing the realism of a quantity that is known a priori to be biased. After excluding the intercept from H0, the resulting comparison is equivalent to the routine procedure of subtracting the mean of each time series prior to analysis and then assessing the stochastic similarity of the anomalies (provided the degrees of freedom are adjusted appropriately).
If H0 is rejected, then a natural question is how individual components of the VARX models contribute to this difference. In the remainder of this section, we describe our procedure for testing and diagnosing differences in VARX model components. The formal justification of this procedure is given in the Appendices.
Hypothesis H0 is tested using a likelihood ratio test. The exact sampling distribution of the test statistic is not available in standard software packages, so we develop a Monte Carlo technique to estimate the associated significance thresholds. Asymptotic theory predicts that the test statistic will have a chi-squared distribution in the limit of a large sample size. However, known biases in maximum likelihood estimates are found to produce serious biases in multivariate problems. Fortunately, a simple bias correction virtually eliminates this bias for the data considered in this paper. The resulting bias-corrected test statistic is called a deviance. For our data, significance thresholds computed from the chi-squared distribution and from the Monte Carlo technique are very close. For other data sets, particularly those with small or disparate sample sizes, the chi-squared distribution may not be adequate and the Monte Carlo technique may be required.
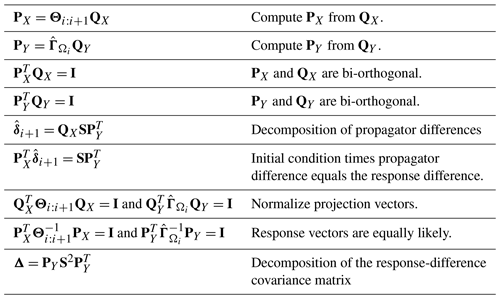
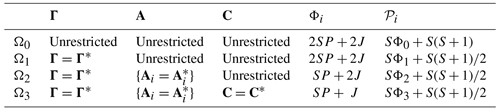
If H0 is rejected, then we conclude that one or more of the VARX parameters differ, but which ones? A natural question is whether the difference in VARX models is due to differences in noise, differences in AR parameters, or differences in annual cycle parameters. This question is addressed by the following stepwise procedure: first test for differences in noise parameters, and if those are not significant, then test for differences in AR parameters, and if those are not significant, then test for differences in annual cycle parameters. This hierarchy of hypotheses is summarized in Table 1. Further details about the stepwise procedure, including the family-wise error rate and the reasons for choosing the particular order, are given in Sect. B7.
Table 1Summary of hypotheses in the stepwise test procedure for VARX(p) models. Parameters Φi and 𝒫i in the last two columns indicate the number of predictors and the number of estimated parameters under Hypothesis Ωi, respectively.
The statistic for testing Ωi versus Ωi+1 is called sub-deviance . Sub-deviance is small when the relevant parameter differences are small and large when the parameter differences are large. The associated significance thresholds are computed using a Monte Carlo technique (see Sect. B6). For our data, these thresholds are consistent with asymptotic theory, which indicates that, if Ωi+1 is true, then asymptotically is chi-squared distributed with degrees of freedom, which we denote as
where 𝒫i is the number of parameters estimated under Ωi and specified in Table 1. The stepwise procedure halts at the first significant sub-deviance.
The deviance for testing the equality of all VARX parameters is the sum of the sub-deviances:
Decomposition (5) is satisfied regardless of the status of the hypotheses in Table 1. Using these terms to quantify the fraction of deviance due to differences in noise, AR, or annual cycle parameters can be subtle. In particular, the largest term may not dictate significance. For instance, one term on the right-hand side might explain 90 % of the deviance and yet still be insignificant, whereas another term explains only 5 % of the deviance yet is significant. This issue should be kept in mind when quantifying the contribution of particular components of the VARX model to the total deviance.
The above procedure might attribute differences to a single part of the VARX model, but that part still involves many parameters, which hinders interpretation. To further isolate VARX model differences, we seek the linear combination of variables that maximizes the appropriate sub-deviance . A procedure called covariance discriminant analysis (CDA DelSole and Tippett, 2022a) can find this combination and leads to a decomposition that can be viewed as a generalization of principal component analysis. Specifically, CDA decomposes each into S components such that the first maximizes , the second maximizes subject to being uncorrelated with the first, and so on.
In the case of , the interpretation of discriminant components is clear-cut: these components identify the spatial structures that exhibit the greatest discrepancies in whitened variances between the two data sets (see DelSole and Tippett, 2022a). For and , however, the connection between these components and differences in VARX parameters is less apparent. In the Appendix, we clarify this connection by showing that sub-deviances can be written as an explicit function of differences in estimated regression parameters (see Sect. B8). To our knowledge, this result has not been derived previously. To describe this relation, let the matrix denote the difference between estimated regression parameters relevant to comparing hypotheses Ωi and Ωi+1. For instance, to compare the Ω1 and Ω2 of a VARX(1) model, . Then, the Appendix shows that the sub-deviance can be written as
where is a linear transformation of (given explicitly in Eq. B34). A singular value decomposition (SVD) of then yields the desired decomposition of . From the SVD, one can either examine differences in regression parameters directly or examine their impact on the model. The latter is often more informative. For instance, when diagnosing differences in AR parameters, the decomposition can be expressed as
where is the difference in AR parameters under Ω1, S is a diagonal matrix, and the columns of PX and PY define initial condition and response patterns, respectively (all the terms are defined precisely in Sect. B8). This decomposition has the following form: the initial condition (PX) multiplied by the difference in AR parameters () equals the difference in the response (PYS). This decomposition can be interpreted as finding the initial condition, among all equally likely initial states, that maximizes the disparity in one-step responses. This decomposition is similar to the “optimal initial condition” discussed in previous studies (e.g., Penland and Sardeshmukh, 1995; Alexander et al., 2008), except here the initial and final vectors are constrained by norms associated with the deviance measure.
To diagnose differences in annual cycle parameters, the above decomposition is not very meaningful because the predictor is a fixed function of time (e.g., sinusoidal functions of time) rather than a random vector. In this case, we simply propagate parameter differences into the time domain by multiplying by the associated predictors X, yielding a decomposition that is similar to principal component analysis in the form
In this section, we apply the above method to compare the variability of monthly mean North Atlantic sea surface temperature (SST) between dynamical models and observations. We analyze the Atlantic basin over 0–60∘ N, where the northern boundary was chosen to avoid regions of sea ice. For observations, we use version 5 of the Extended Reconstructed Sea Surface Temperature (ERSSTv5) of Huang et al. (2017). The earlier portion of this data set has sparse spatial coverage, so we only analyze the more recent 50-year period 1969–2018.
For model simulations, we use data from Phase 5 of the Coupled Model Intercomparison Project (CMIP5; Taylor et al., 2012). Specifically, we use pre-industrial control runs, which are simulations without year-to-year changes in external forcing like atmospheric composition or solar insolation. The associated initial condition is effectively random, and hence these simulations are independent, consistent with our assumptions in Eqs. (1) and (2). We select models only if their control runs are at least 500 years long. This criterion selects 36 models from the CMIP5 data set. We extract the last 50 years of each simulation. We examine the last two 25-year segments of each simulation and compare them with each other and with 25-year segments from observational data. While our test can accommodate time series of varying lengths, we found that a duration of 25 years is sufficient for detecting differences effectively.
In general, the above time series contain secular trends: observations contain a global warming signal and control runs contain model drift. These secular trends will be removed by regressing out a polynomial in time. It is important to remove the same-order polynomial from both observations and models; otherwise, a difference could arise simply as an artifact of processing the two time series differently. It is known that greenhouse gas concentrations rise exponentially over our analysis period (1969–2018), so if we were to remove only a linear trend and then find a difference, that difference could be attributed to quadratic growth in the observations that is missing from control runs. In general, leaving any kind of forced variability in time series leads to problems of interpretation since our VARX model does not account for secular forcing. On the other hand, over-removal of low-frequency internal variability poses a lesser issue. While the resulting analysis would not relate to low-frequency variability (since it was removed), the conclusions regarding higher-frequency variability would still retain their validity. Hence, it is generally preferable to err on the side of removing a higher-order polynomial than a lower-order one. For the results presented in the figures below, a second-order polynomial in time was removed over the period 1969–2018. However, removing third-, fourth-, or higher-order polynomials removes additional low-frequency variability from the time series but does not alter any of our main conclusions regarding significant differences between observations and CMIP5 models.
For the annual cycle term, we include five annual harmonics encompassing all harmonics up to the Nyquist frequency. This choice is motivated by the same reasoning as discussed above for secular trends, i.e., that it is preferable to include more harmonics rather than fewer. If we were to select an insufficient number of harmonics, then unaccounted-for periodic signals would be misattributed as internal variability in the VARX model, leading to erroneous conclusions. On the other hand, if an excessive number of harmonics is chosen, the VARX model may become overfitted, but this potential overfitting is accounted for in the sampling distribution. By definition, overfitting implies the inclusion of predictors with vanishing regression coefficients, but the sampling distribution is independent of the specific values of the regression coefficients and therefore encompasses cases where the coefficients are zero. The primary drawback of overfitting is a reduction in statistical power. However, in our specific application, low statistical power is not a concern: our method demonstrates high effectiveness in detecting differences when they truly exist. Therefore, the negative consequences associated with overfitting, such as diminished statistical power, are not a significant issue in this context.
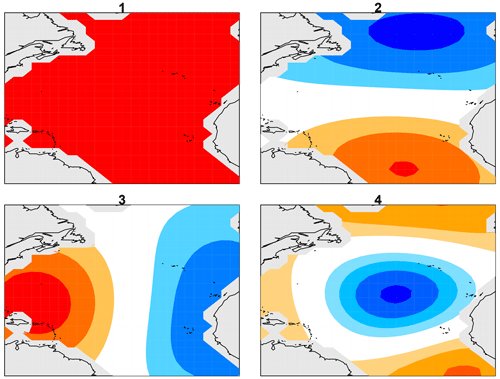
Figure 1Laplacian eigenvectors 1, 2, 3, and 4 over the North Atlantic between the Equator and 60∘ N, where dark red and dark blue indicate extreme positive and negative values, respectively.
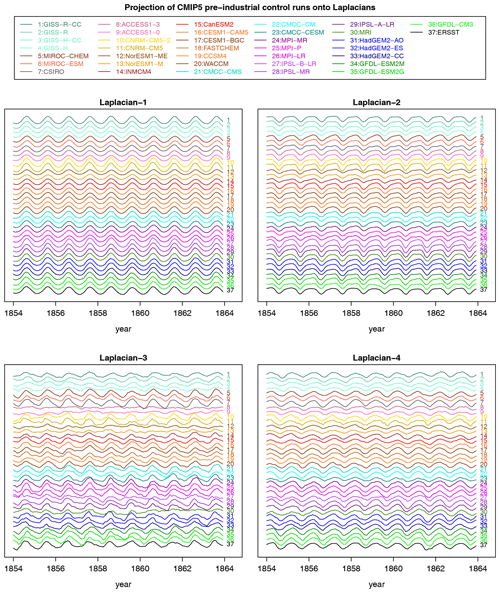
Figure 2Monthly time series of Laplacians 1–4 in observations (black curve at the bottom labeled “37”) and in CMIP5 models over a 10-year period. Each time series is offset by the same constant. No re-scaling is performed. Each time series is computed by projecting data onto the appropriate Laplacian over the Atlantic domain. The year on the x axis corresponds to the observational data set ERSSTv5 but otherwise is a reference label for the pre-industrial control simulations.
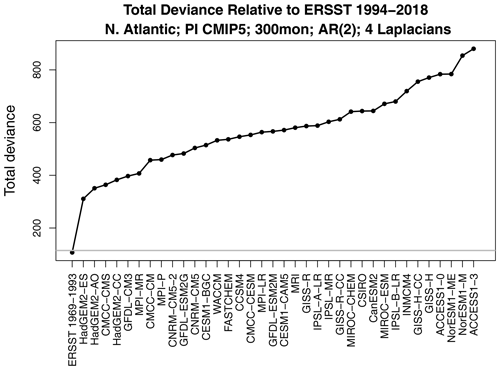
Figure 3The total deviance between ERSSTv5 1994–2018 and each CMIP5 model. The horizontal gray line shows the 1 % significance threshold. Also shown is the deviance between ERSSTv5 for the two periods 1969–1993 and 1994–2018 (first x-tick mark on the left).
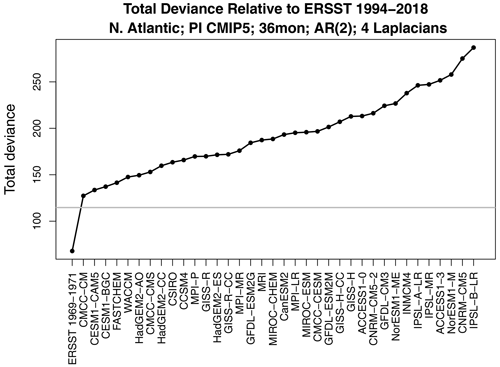
Figure 4Same as Fig. 3 but using only 3 years of data from each CMIP5 model. Also shown is the deviance between ERSSTv5 for the two periods 1969–1971 (3 years) and 1994–2018 (first x-tick mark on the left).
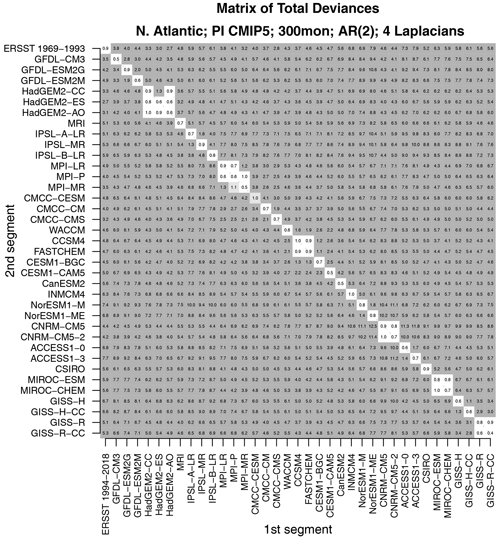
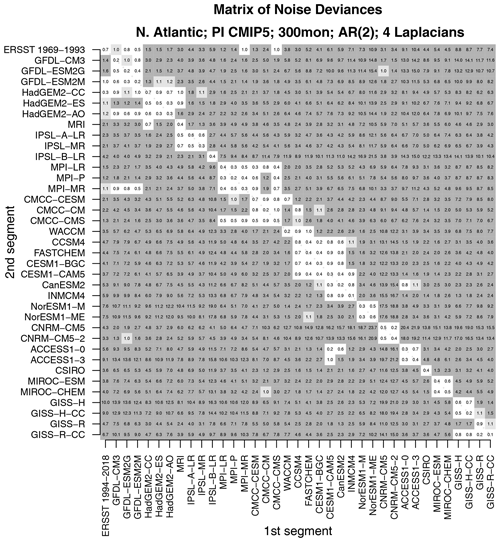
Figure 5The total deviance between each 25-year segment from CMIP5 models and observations to an independent 25-year segment from CMIP5 models and observations. The deviance is normalized by the 1 % significance threshold. Values that are insignificant, significant at the 1 % level, and significant at the 0.2 % level are indicated by no shading, light gray shading, and dark gray shading, respectively.
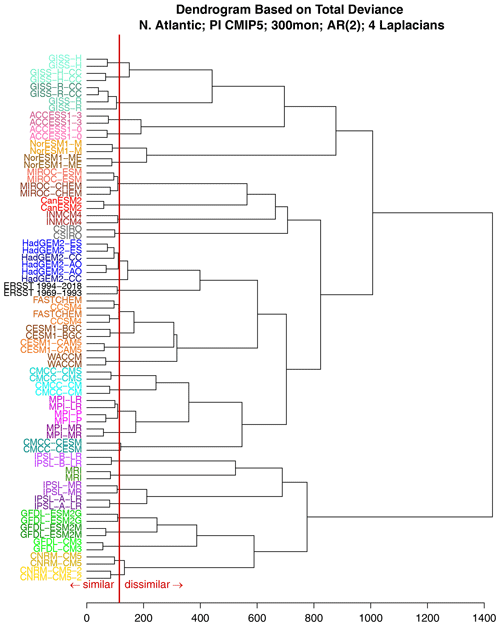
Figure 6A dendrogram based on the total deviance of monthly mean North Atlantic SST. The x axis is the deviance. Larger deviance values imply stronger dissimilarities between the VARX models. All the time series are 25 years in length. Two separate 25-year segments from CMIP5 models and from observations (ERSSTv5) are included in the cluster analysis. The vertical red line shows the 1 % significance level.
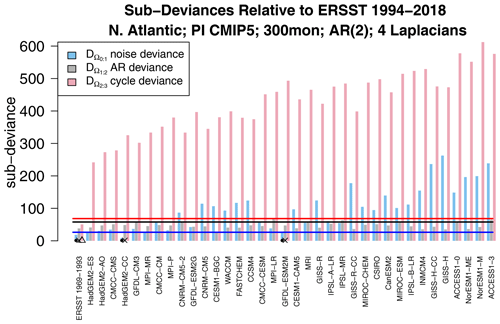
Figure 7Decomposition of each deviance in Fig. 3 into differences in noise parameters (blue), differences in AR parameters (green), and differences in annual cycle parameters (red). The associated 1 % significance thresholds are indicated by the blue, green, and red horizontal lines, and sub-deviances that are insignificant according to the stepwise procedure are indicated by the dot, cross, and triangle, respectively.
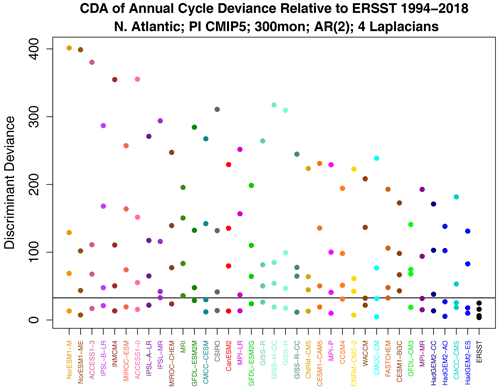
Figure 8Decomposition of annual cycle deviance by discriminant analysis. The horizontal gray line is the 1 % significance threshold for the maximum deviance, computed as described at the end of Sect. B6.
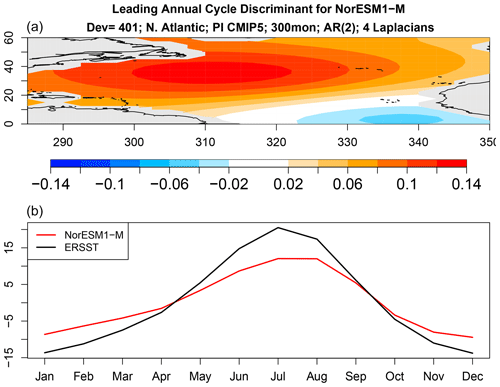
Figure 9The spatial pattern (a) and time series (b) of the leading discriminant of the annual cycle deviance for NorESM1-M.
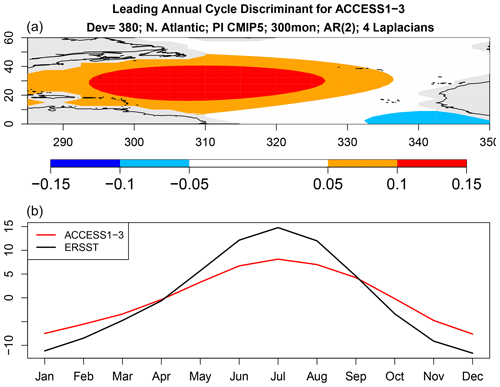
Figure 10Same as Fig. 9 but for ACCESS1-3.
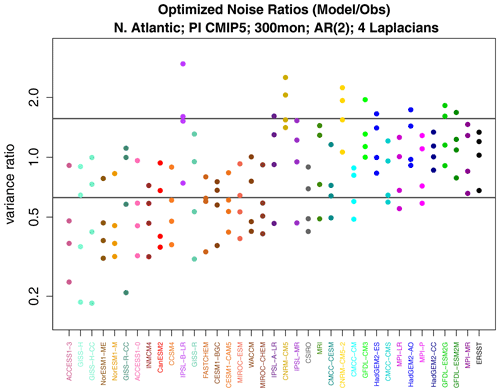
Figure 11Variance ratios from discriminant analysis of noise variance, where the ratio is CMIP5 model noise variance over observation noise variance. The two horizontal gray lines indicate the upper and lower 0.5 % significance levels.
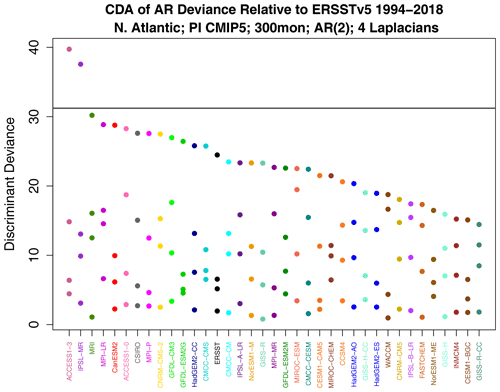
Figure 13Decomposed AR deviance from discriminant analysis. The horizontal gray line indicates the 1 % significance threshold. Models on the x axis have been ordered by leading deviance.
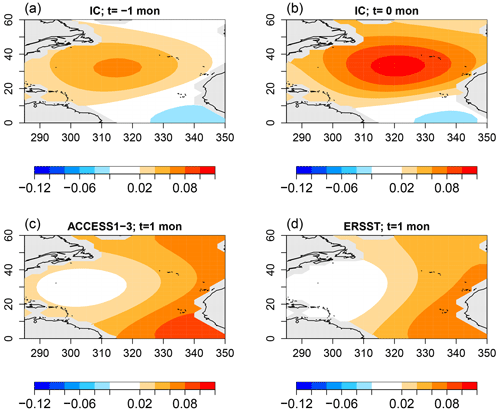
Figure 14The optimal initial condition of VARX(2) models (a, b) that maximizes the difference in response between ACCESS1-3 and ERSSTv5 and the corresponding response in ACCESS1-3 (c) and ERSSTv5 (d).
As in many climate applications, the spatial dimension in our data set exceeds the time dimension, leading to an underdetermined estimation problem. Although several regularization approaches are available, many of these have no rigorous hypothesis test framework. Here, we regularize the problem by reducing the spatial dimension, which retains the regression model framework. The question arises as to which low-dimensional space should be selected. Our choice is guided by the fact that numerical solutions have less reliability with a decreasing spatial scale, with the least reliable results at the grid-point scale. These considerations suggest that models are most reliable on the largest spatial scales, and therefore a feature space should be chosen to emphasize large spatial scales. A common approach is to use empirical orthogonal functions (EOFs), but EOFs depend on data and therefore raise the question as to which data should be used to derive them. Also, there is no guarantee that the EOFs will be strictly large scale. Furthermore, because EOFs depend on data, their use leads to biases and random fluctuations that are not straightforward to take into account in the final statistical estimate. An attractive alternative basis set that avoids these issues and satisfies the above requirement contains the leading eigenvectors of Laplace's equation. These vectors form an orthogonal set of spatial patterns ordered by the decreasing spatial scale. Familiar examples of Laplacian eigenvectors include Fourier series and spherical harmonics. The algorithm of DelSole and Tippett (2015) was used to compute the Laplacian eigenvectors over the Atlantic domain. The first four eigenvectors are shown in Fig. 1. The first Laplacian eigenvector is spatially uniform, and therefore projecting data onto the first vector yields the spatial mean. The spatial mean of Atlantic SSTs after removal of human-caused variations is often called the Atlantic Multidecadal Variability (AMV) index. The second and third eigenvectors are dipoles measuring the north–south and east–west gradients across the basin. Subsequent vectors capture smaller-scale spatial structures. Compared to the usual choice of EOFs, Laplacian eigenvectors are particularly attractive because the first component is the AMV index, a natural climate index, and the set depends only on the domain geometry and hence is independent of data and independent of the model.
How many Laplacian eigenvectors should be chosen? In this study, our goal is to compare variability, not to make predictions. Accordingly, our primary concern is to ensure that the VARX(p) model is adequate. In practice, adequacy means confirming that the residuals of the estimated VARX(p) model are indistinguishable from white noise. The multivariate Ljung–Box test was performed on the residuals of the VARX(p) models to check for whiteness. For p=1, over half the VARX models exhibited significant non-whiteness at the 5 % level, regardless of the number of eigenvectors S. For p=2, only 16 % of the VARX models exhibited significant non-whiteness for S=4, with this percentage increasing for S>4. After a Bonferroni correction, significant non-whiteness was detected in only 1 out of 37 VARX models for S=4, suggesting that the VARX(2) model with S=4 and 5 annual harmonics is adequate.
For reference, 10-year segments of time series for the first four Laplacian eigenvectors are shown in Fig. 2. The strong periodicity seen in Laplacian-1 reflects the fact that 99 % of the variance is explained by the annual cycle. The strongest discrepancies between time series are seen in Laplacian-3, but this assessment is merely visual and subjective.
The total deviance between ERSSTv5 1994–2018 and each CMIP5 model is shown in Fig. 3. The corresponding 1 % significance threshold computed from Eq. (4) is indicated by the horizontal gray line. The first x-tick mark shows the deviance between ERSSTv5 for the two periods 1969–1993 and 1994–2018, which falls below the gray line, indicating no significant difference in stochastic processes between the two observational periods. In contrast, the deviance between observations and each CMIP5 model lies well above the significance threshold, indicating strong differences between stochastic processes. The same conclusion is reached when the reference period is switched from 1994–2018 to 1969–1993 (not shown).
The above examples are based on using 25 years of data for both climate models and observations. Adding more data merely makes the differences detected here even more significant. An interesting question is whether differences can be detected using shorter samples from climate models. Recomputing deviances using only 3 years of data (but still using 25 years of observational data) yields the results shown in Fig. 4. Remarkably, differences can still be detected, as indicated by the fact that all the deviances lie above the significance threshold. The same conclusion holds if we swap these numbers, i.e., use only 3 years of observations and 25 years of model data (not shown).
Two questions arise naturally here. First, do CMIP5 models differ from observations in a common way? This question can be addressed by comparing one model to another – a small deviance would indicate that the two models are similar and therefore differ from observations in a common way. Second, would the test correctly indicate that two time series from the same CMIP5 model are stochastically similar? Intuitively, data generated by the same model ought to be stochastically similar. However, this outcome is not assured. For instance, CMIP5 models are nonlinear and high-dimensional. There is no guarantee that variability from such models can be captured by a low-dimensional linear model. Also, our test assumes that sample sizes are sufficiently large to invoke a linear regression framework for testing hypotheses. Twenty-five years of data might not satisfy this requirement. These latter questions can be addressed by confirming that independent segments from the same CMIP5 model are stochastically indistinguishable. Both questions can be addressed by comparing one 25-year segment with a separate 25-year segment for all possible pairs of CMIP5 models and observations. The result of comparing all possible pairs is summarized in the matrix shown in Fig. 5.
As can be seen, values along the diagonal of this matrix are insignificant. Diagonal elements correspond to comparing time series from the same model or from the same observational data set. Thus, this test indicates that time series from the same source are stochastically indistinguishable, confirming that the test performs as expected. Additional insignificant deviances are found in diagonal blocks. For instance, the 2×2 block in the upper-left corner indicates that the two CNRM models are stochastically indistinguishable. The next blocks include the HadGEM2, MPI, NCAR, MIROC, and GISS models. In other words, models from the same center can be stochastically indistinguishable. Other than those cases, all other model–model deviances are significant. In short, models differ not only from observations, but also from each other, unless they come from the same center.
Although all the models are different, some models are more similar to each other than to others. To identify clusters, we compute a dendrogram from these deviances. The dendrogram is computed in the following way. First, each element is assigned to its own cluster. Next, the pair with the smallest deviance is clustered together using a “leaf” whose edge aligns with the deviance indicated on the x axis. Next, the pairs with the next smallest deviance are clustered together in the same way. Clusters themselves are joined to other elements or clusters using the complete-linkage rule, whereby the length of the leaf equals the maximum deviance among all pairs of members of the cluster. Each new cluster creates a new leaf further to the left corresponding to larger deviances. The resulting dendrogram is shown in Fig. 6.
Remarkably, the majority of model names on the left-hand side of the dendrogram are grouped into consecutive pairs, indicating a consistent pairing of time series originating from the same source. This suggests that each CMIP5 model simulation possesses a distinct “fingerprint” that can be quantified using the deviance measure. However, there are a few exceptions to this pattern, i.e., HadGEM2-CC, FASTCHEM, and CCSM4. These exceptions are paired with other models from the same modeling center, supporting the earlier conclusion that models from the same center may exhibit similarities that make them indistinguishable. Consequently, CMIP5 models from the same modeling center tend to share comparable fingerprints.
Which models are closest to the observations? According to the dendrogram, after clustering the observational time series together, they are subsequently grouped with HadGEM2 models and then with NCAR models (CESM, CCSM, and FASTCHEM). Importantly, the leaves connecting to observations surpass the significance threshold, consistent with the findings shown in Fig. 4. This implies that the HadGEM2 and NCAR models are the closest to the observations, although they still clearly differ from observations.
A natural question is whether the difference in VARX models is due to differences in noise parameters, AR parameters, or annual cycle parameters. This question can be addressed by computing the decomposition in Eq. (5). The results are shown in Fig. 7. We see immediately that the largest source of deviance is the annual cycle. In fact, if the annual cycle deviance were computed for all possible pairs, virtually the same structure as in Fig. 5 would emerge (not shown). This confirms that the annual cycle of any CMIP5 model differs not only from observations, but also from other models from different modeling centers. In other words, the annual cycle is also an effective fingerprint.
The above results show that the annual cycle in each CMIP5 model differs from observations and from other CMIP5 models, but they do not tell us how they differ. We can anticipate from Fig. 5 that there is no common difference, so any description of model error will be model-dependent. Nevertheless, even for a single model, isolating key differences is difficult because the annual cycle lies in a high-dimensional space. One could study plots of annual cycle differences, but eye-catching features may not be the features that contribute most to annual cycle deviance. This is where covariance discriminant analysis proves useful. Discriminant analysis decomposes sub-deviance into uncorrelated components ordered such that the first explains the most sub-deviance, the second explains the most sub-deviance subject to being uncorrelated with the first, and so on. If a few components explain all the sub-deviance, then discriminant analysis will find them. The result of applying discriminant analysis to decompose annual cycle deviance is shown in Fig. 8. Since the VARX model is based on four Laplacian eigenvectors, discriminant analysis decomposes sub-deviance into four components per model. The leading discriminant deviance is significant for all the CMIP5 models. In some models, the leading discriminant deviance exceeds the second by a factor of 3 or more (e.g., NorESM1 models and ACCESS1-3). The associated time series and pattern from decomposition (7) for NorESM1-M and ACCESS1-3 are shown in Figs. 9 and 10. Time series for observations and CMIP5 models separately are computed as described in Sect. B8 (see Eq. B41). The two figures suggest that the two CMIP models underestimate the winter and summer peaks in the Gulf Stream regions.
Sub-deviances that are insignificant according to the stepwise procedure are indicated by the dot, cross, and triangle for differences in noise, AR parameters, and annual cycle parameters, respectively. Only two models have insignificant differences in noise and AR parameters: HadGEM2-CC and GFDL-ESM2M. This means that the internal variability of these models is consistent with observations. To be fair, the noise and AR parameters of some other CMIP5 models are only marginally inconsistent with observations (e.g., the HadGEM2, MPI, and CMCC models). Also, if the observational reference is changed to the 1969–1993 period, then HadGEM2 is no longer consistent (not shown). In all the cases, the annual cycle of each CMIP5 model differs significantly from observations.
For some CMIP5 models, differences in noise parameters explain a large fraction of the total deviance. To assess whether these models exhibit common discrepancies in whitened variance, we calculate the noise deviance between all possible combinations of models and observations. The corresponding results are shown in Fig. 12. Relative to the total deviance shown in Fig. 5, noise deviance exhibits more cases of indistinguishable models. Still, many of these cases involve models originating from the same research center. Furthermore, each model remains distinctly different from the majority of other models, indicating that whitened variance alone can serve as a distinguishing feature or fingerprint. It is rather remarkable that, even after removing serial correlations, annual cycles, and trends, the remaining variability contains sufficient information to differentiate a model from most other models. Recall that the stepwise procedure begins by comparing whitened variances. This result shows that this first step already has large discriminative power even before differences in AR or annual cycle parameters are considered.
We applied covariance discriminant analysis to determine whether a few spatial structures can explain the noise deviance. The resulting discriminant ratios are shown in Fig. 11. Ratios are shown instead of deviances because ratios differentiate the direction of variance difference, whereas deviance is insensitive to direction as both smaller and larger whitened variances contribute to positive deviances. Each model yields four discriminant ratios since the VARX is based on four Laplacian eigenvectors. The strongest separations from one are those below one, indicating that CMIP5 models exhibit too little whitened variance compared to observations. Examination of the leading discriminant patterns for noise deviance across CMIP5 models (not shown) reveals no common bias structure, as one would anticipate from Fig. 12.
According to Fig. 7, the sub-deviance of AR parameter differences is relatively small. Nevertheless, for completeness, we show the associated discriminants. The discriminant deviances are shown in Fig. 13. As expected, most leading values are insignificant. The largest leading value occurs for ACCESS1-3. The associated AR parameter difference can be expressed as an initial value problem using Eq. (6). Note that this identifies the initial condition of a VARX(2) model, so the initial condition is defined by two SST patterns, one at the initial month and one at the antecedent month. The resulting patterns are shown in Fig. 14. The figure suggests that, for this initial condition, ACCESS1-3 responds with a stronger amplitude than observations, although the amplitude discrepancy is modest, consistent with the modest deviance level.
This paper presents a methodology for determining whether two vector time series originate from the same stochastic process. Such a procedure can be used to address various climate-related questions, including assessing the realism of climate simulations and quantifying changes in climate variability over time. The stochastic process under consideration is assumed to be a vector autoregressive model with exogenous variables, referred to as VARX. In this study, the exogenous variable represents annual cycles in the mean. However, in other applications, it could capture nonstationary signals such as diurnal cycles, secular changes due to solar variability, volcanic eruptions, or human-induced climate change. This paper derives a likelihood ratio test for determining the equality of VARX parameters. Additionally, an associated stepwise procedure is developed to determine the equality of noise parameters, autoregressive parameters, and annual cycle parameters. The resulting procedure is not limited to specific stochastic models employed in this study. Rather, the procedure is general and can be applied to a broader class of models, including non-periodic exogenous variables. Thus, these procedures provide a comprehensive framework for analyzing and comparing different aspects of climate time series.
Derivation of the above procedure follows an approach that is similar to the univariate case, but it is extended here to encompass multivariate applications. This extension necessitates the incorporation of bias corrections and the utilization of a Monte Carlo technique to estimate significance thresholds accurately. The Monte Carlo algorithm developed here is particularly efficient in that it uses eigenvalue methods to evaluate the ratio of determinants and avoids solving regression problems by sampling directly from the Wishart distribution. Discriminant techniques are employed to compress differences between VARX models into the minimal number of components, facilitating a more concise description. While similar techniques were introduced in previous parts of this paper series, this paper generalizes them to multivariate situations and to accommodate an arbitrary number of steps in the stepwise procedure. Consequently, the procedure and associated codes for this test supersede those discussed in earlier parts of the series, offering an improved and more comprehensive approach.
The above procedure was implemented to compare monthly mean North Atlantic sea surface temperatures between CMIP5 models and observational data, taking into account their respective annual cycles. The analysis focused on the variability projected onto the first four Laplacian eigenvectors over the North Atlantic basin, which highlight the largest spatial scales within the region. To ensure that the residuals exhibited properties akin to white noise, a VARX model of at least second order was required for most CMIP5 models. The test results indicated that not only do CMIP5 models differ stochastically from the observational data, but that they also display variations among themselves, except when models originate from the same modeling center. Differences among CMIP5 models are distinctive enough to serve as a fingerprint that differentiates a given model from any other model and from observational data.
The primary source of deviance from observations is disparities in annual cycles. To gain insight into the characteristics of these disparities, covariance discriminant analysis was employed to decompose deviance associated with annual cycles into uncorrelated components, ordered such that the first explains the largest portion of annual cycle deviance, the second explains the most deviance after the first has been removed, and so on. For certain CMIP5 models, the leading discriminant accounts for several times more annual cycle deviance than subsequent components. Specific examples of these leading discriminants were presented.
Although differences in annual cycles dominated the total deviance, differences in whitened variance were also significant across the majority of the models. Discriminant analysis revealed that most CMIP5 models underestimate whitened variance, with some models falling short by a factor of 5. A few models were found to overestimate whitened variance by a factor of 2 or more. The collective differences in whitened variances and covariances between Laplacian eigenvectors were sufficiently unique to serve as a secondary fingerprint. It is remarkable that such distinctive identifying information is encapsulated in time series even after removing serial correlations, annual cycles, and all other nonstationary signals such as trends.
Differences in autoregressive parameters accounted for only a minor portion of the overall deviance. Approximately two models displayed noise and AR parameters that aligned with observations, suggesting that their internal variability was realistically represented. However, all the models exhibited unrealistic annual cycles despite this positive characteristic.
The method discussed in this paper can analyze only relatively low-dimensional systems (for instance, the VARX model used to compare North Atlantic variability examined only four Laplacian eigenfunctions). It may be possible to combine some aspects of this approach with machine learning methods to greatly expand the number of variables that can be compared.
A standard method for testing hypotheses in VAR models is the maximum likelihood method (Brockwell and Davis, 1991; Box et al., 2008; Lütkepohl, 2005). The extension of this method to VARX models is straightforward. Specifically, the likelihood of Eq. (1) is
where LP represents terms that depend on the first p values of the process, and
Similarly, the likelihood of Eq. (2) is
where
Since {yt} and are independent, the likelihood of both models is the product of the two individual likelihoods:
For the alternative hypotheses listed in Table 1, the likelihood is obtained by constraining the population parameters as indicated in Table 1. As an example, the likelihood under Ω3 is
where and take on the values
Maximum likelihood estimates are obtained by finding the parameters that maximize the likelihood in question. Unfortunately, this procedure is complicated by the terms Lp and , which depend nonlinearly on the population parameters. However, for large N′ and , the contribution of Lp and to the likelihood is small in comparison to other terms. A common approximation is to ignore Lp and when computing the maximum likelihood estimates, which is tantamount to maximizing the conditional likelihood and leads to the familiar least-squares estimates (Box et al., 2008, Sects. 7.1.2 and A7.4). Moreover, for asymptotically large sample sizes, the sampling distributions converge to those associated with linear regression theory. For example, see theorem 8.1.2 and Sect. 8.9 of Brockwell and Davis (1991) and Appendix A7.5 of Box et al. (2008) for the univariate case as well as Sect. 3.4 of Lütkepohl (2005) for the multivariate case. Accordingly, we assume that sample sizes are sufficiently large to justify asymptotic theory and therefore justify using linear regression theory to test hypotheses in VARX models. The problem of testing differences in VARX models is then addressed by appropriately mapping the parameters of the VARX model to the parameters of the regression model. This mapping can be performed in the following way. First, define
where L and U denote the lower and upper limits of a time series, respectively, with L<U. Then, the N′ samples from model (1) are related as
where
and
In the context of comparing annual cycles,
where
Similarly, in an obvious notation,
In this way, the problem of comparing VARX models (1) and (2) is re-parameterized as the problem of comparing Eqs. (A3) and (A4). In the next section, we describe the procedure for comparing Eqs. (A3) and (A4). For this description, it proves convenient to define
B1 Regression model framework
Although our goal is to describe the procedure for comparing the regression models (A3) and (A4), it turns out to be more efficient to describe the procedure in the context of the more general problem of comparing the models:
where Xk and are predictor matrices, each having linearly independent columns; Bk and are regression coefficients; and E and E* are independent random matrices whose rows are drawn from a multivariate normal distribution with zero mean and covariance matrices Γ and Γ*, respectively. These terms have the following dimensions:
where , and N and N* are defined in Eq. (A5). S is the number of variables (i.e., the dimension of yt in Eq. 1). are the number of predictors in .
Samples from VARX(p) models (1) and (2) can be written in the regression model forms (B1) and (B2) as discussed in Appendix A. The partitioning of parameters in models (1) and (2) into the Bi matrices in Eqs. (B1) and (B2) is highly adaptable and allows for considerable flexibility. To test the hypotheses in Table 1, B2 would be identified with the AR parameters , B3 would be identified with the exogenous parameters C, and B4 would be identified with the intercept. It is a matter of choice whether to test the equality of B4. In Sect. 3, equality of climatological means was not tested, and hence the equality of B4 was not tested. Under this identification, K2=pS, K3=J, and K4=1. The identification of starred variables follows the same pattern. The null hypothesis H0 in Eq. (3) corresponds to the hypothesis that for and .
B2 Stepwise testing procedure
If H0 is rejected, then one or more of the equalities in Eq. (3) is false, but which ones? To address this question, we test hypotheses about subsets of parameters. The hypothesis is tested first for reasons discussed in DelSole and Tippett (2022b). Then, hypotheses about other parameter subsets are tested. Seber (2015) describes an elegant procedure for testing hypotheses in a stepwise manner. This procedure was adopted in DelSole and Tippett (2022b) and will be generalized further in this Appendix to include multivariate models. The least restrictive hypothesis is denoted as Ω0, and subsequent hypotheses are nested as . The first few hypotheses are indicated in Table B1. Note that Hypothesis H0 is not Ω0. Also, model (B1) deliberately excludes X1 and B1, so that hypotheses in Table B1 correspond to restricting B2,B3, and B4, respectively. Had the subscripts of B started at one, the resulting subscripts on Ω and B would have differed by one, leading to an awkward notation.
Table B1Summary of the first few hypotheses in the stepwise procedure. Parameters Φi and 𝒫i in the last two columns indicate the number of predictors and the number of estimated parameters under Hypothesis Ωi, respectively.
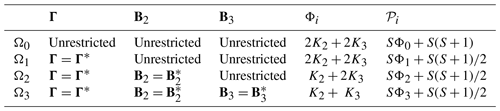
A key quantity in the stepwise procedure is the number of parameters estimated under the ith hypothesis, denoted as 𝒫i. Table B1 summarizes the number of parameters 𝒫i and the number of predictors Φi associated with each hypothesis. For instance, the population parameters in Eq. (B1) are . Each Bi contains Ki predictors and therefore SKi parameters, and Γ contains independent parameters. Model (B2) contains the same number of parameters. Therefore, the total number of parameters estimated under Ω0 is
Under Ω1, the population parameters are . In particular, there is only one noise covariance matrix, so
As the stepwise procedure marches from Ωi to Ωi+1, there are Ki+1 fewer predictors estimated. Thus, in general, the number of parameters estimated under Ωi for is
where H(x) is a step function, which equals 0 when x≤0 and equals 1 when x>0 (note that H(0)=0).
B3 Equality of noise covariance matrices
The initial hypothesis in the stepwise test procedure is Ω1, which asserts the equality of noise covariance matrices. This test shares similarities with the conventional test for equality of covariance matrices (e.g., Anderson, 1984, Chap. 10). However, certain adjustments are made, as discussed in more detail below, including an adjustment in the degrees of freedom to accommodate regression and the implementation of a bias correction. We start by expressing Eq. (B1) in the following form:
where
By standard theorems, the maximum likelihood estimate of 𝔹 is
the maximum likelihood estimate of Γ is
and the distribution of is
where the right-hand side denotes an S-dimensional Wishart distribution with ν degrees of freedom and covariance matrix Γ. The degrees of freedom for model (B1) is
Analogous results hold for model (B2), where the associated estimates are distinguished by asterisks. In particular, the estimated noise covariance matrix has distribution
where
The likelihood ratio for testing Ω1 versus Ω0 is
where denotes the determinant of a matrix and is the maximum likelihood estimate of Γ under Ω1, i.e.,
The corresponding sub-deviance statistic is therefore
We have found that the sampling distribution of the above statistic is grossly inconsistent with asymptotic theory, particularly for disparate N and N*. This inconsistency is most likely due to the well-known fact that the maximum likelihood estimate in Eq. (B4) is biased. In principle, an unbiased estimate could be obtained simply by replacing N in Eq. (B4) with ν. Previous studies have shown that replacing maximum likelihood estimates with unbiased estimates yields a likelihood ratio for finite samples that is more consistent with asymptotic sampling theory (Cordeiro and Cribari-Neto, 2014). This correction becomes even more critical with larger S. A suitable bias-corrected deviance can be obtained by replacing N and N* with the degrees of freedom ν and ν*, replacing the maximum likelihood estimate of Γ in Eq. (B4) with the unbiased estimate
and making a similar replacement of the maximum likelihood estimate with the unbiased estimate . Furthermore, an unbiased estimate of Γ under Ω1 is
The resulting bias-corrected sub-deviance is
B4 Equality of regression parameters
Procedures for testing hypotheses are standard (e.g., Anderson, 1984, Sect. 8.3–8.4) and are sometimes referred to as testing a subset of explanatory variables (Fujikoshi et al., 2010, p191). The procedure we follow is similar, except that we employ estimates of noise covariance matrices that connect appropriately to the bias-corrected estimates defined in Sect. B3.
Under hypotheses , models (B1) and (B2) have a common noise covariance matrix, and thus the models can be combined in the form , where
and and are defined based on the hypothesis in question. For instance, for κ=3, the identifications for Ω1,Ω2, and Ω3 would be
and
The maximum likelihood estimate of Γ under Ωi is
where
The corresponding (maximum likelihood) sub-deviance for testing Ωi+1 against Ωi is
As in Sect. B3, we modify the noise covariance matrix estimate and the sub-deviance to satisfy certain properties. Accordingly, consider the modified noise covariance matrix estimate
and the corresponding sub-deviance
where the coefficients ai and bi are to be determined. In general, sub-deviance should vanish when the regression coefficients under Ωi and Ωi+1 are identical. In this case, the sum square errors are identical, i.e., , and the sub-deviance is
For sub-deviance to vanish for all 𝔼i, we must have , which in turn implies . Furthermore, the case i=1 is known from Sect. B3. In particular, Eq. (B10) implies . Substituting this into Eq. (B13) yields
Also, consistency with Eq. (B11) requires , which implies that the sub-deviance for testing Ωi+1 against Ωi is
In general, deviance is the telescoping sum of the appropriate sub-deviances:
Estimate Eq. (B14) is biased for i>1 because the degrees of freedom associated with Ωi is , which differs from for i>1. Despite this bias, we use Eq. (B14) because, as shown above, it ensures that each sub-deviance vanishes when the appropriate regression parameter estimates are equal.
B5 Numerical evaluation of sub-deviances
This section describes the computation of sub-deviances using eigenvalue methods. These methods effectively handle underflow and overflow issues and seamlessly integrate with the diagnostic procedures discussed in Sect. B8.
To evaluate in Eq. (B11), we solve the eigenvalue problem
Let the resulting eigenvalues be denoted as . A classical result (Seber, 2008, Sect. 16.8) is that, since and are positive definite, then and are simultaneously diagonalizable and can be written in the forms
where P is nonsingular and Λ is a diagonal matrix with diagonal elements equal to the eigenvalues of Eq. (B17). Substituting Eq. (B18) into (B11) and using standard properties of determinants to cancel P yields
Similarly, to evaluate in Eq. (B15), one could solve
However, for consistency with the calculations discussed later, we solve
There is no material difference between solving Eqs. (B20) and (B21): each one is solved by the same eigenvector q, and the associated eigenvalue is related as . Let the eigenvalues of Eq. (B21) be . Then
Sub-deviance depends only on the eigenvalues of the relevant matrices. The corresponding eigenvectors are not needed for computing sub-deviance, but we will show in Sect. B8 that they are useful for diagnosing the cause of the deviance.
B6 Significance testing
For sufficiently large N and N*, asymptotic theory (Hogg et al., 2019) implies that
We have checked this distribution against Monte Carlo estimates and found that the above asymptotic distribution is reasonable for our data set, provided that unbiased deviances are used. In contrast, if maximum likelihood estimates are used, then the critical value from asymptotic theory can be grossly inaccurate. Once the bias correction is performed, the Monte Carlo algorithm is not essential for the results described in this paper. However, for other data sets with small or disparate N or N*, the asymptotic distribution may break down and the Monte Carlo algorithm may be required, so we summarize the algorithm here. The Monte Carlo technique proposed here is particularly efficient in that it avoids explicit solution of regression problems and instead samples directly from Wishart distributions.
The distributions of , and are known from standard regression analysis (e.g., see Mardia et al., 1979) and are given by
where 𝒲S(ν,ΓC) denotes an S-dimensional Wishart distribution with ν degrees of freedom and covariance matrix ΓC. Importantly, and appearing in Eq. (B15) are not independent. Rather, and are independent (this is proven in Sect. C). Accordingly, it proves convenient to define
and to write Eq. (B15) equivalently as
Section C shows that the distribution of under Ωi+1 is
Importantly, each sub-deviance is invariant to invertible linear transformations of the variables. Therefore, without loss of generality, we may choose ΓC=I. Note that most mathematical packages provide a function to sample directly from the Wishart distribution (e.g., rWishart in R). Sampling directly from the Wishart distribution is much more efficient than repeatedly solving regression problems.
Accordingly, we draw random matrices from
Then, Monte Carlo samples of the sub-deviances are generated as
where the first three terms are
A straightforward Monte Carlo algorithm draws samples from Eqs. (B24) to (B26), computes each sub-deviance in Eq. (B27), and repeats enough times (e.g., 5000) to accurately estimate the 95th percentile of the desired sub-deviance. Here, each sub-deviance is computed using the eigenvalue methods of Sect. B5. Furthermore, individual eigenvalues are retained from the Monte Carlo algorithm, so that they can be used to estimate the sampling distribution of the leading eigenvalues as part of the discriminant analysis discussed in Sect. B8.
B7 Additional comments about the stepwise procedure
Because the hypotheses in Table B1 are nested, there exists a natural stepwise testing procedure. This procedure has been described elegantly by Hogg (1961) and Seber (2015) for univariate regression, which formed the basis of the procedure in DelSole and Tippett (2022b). The procedure for multivariate regression is identical. These steps are described in DelSole and Tippett (2022b) and are repeated below for convenience. First, is tested for significance. If it is significantly large, then we decide that Ω1 is false and stop testing. On the other hand, if is not significant, then is tested for significance. If it is significant, then we decide that Ω1 is true but Ω2 is false and stop testing. On the other hand, if is not significant, then is tested for significance. If it is significant, then we conclude that Ω2 is true but Ω3 is false. If is not significant, then we conclude that no significant difference in VARX models is detected.
Because sub-deviances are stochastically independent, the family-wise error rate associated with multiple testing can be constrained. In this paper, we fix the type-1 error rate of each sub-test to , which gives a 1 % family-wise error rate for the difference-in-VARX model test.
The preferred order of testing the hypotheses listed in Table 1 is discussed in DelSole and Tippett (2022b). This order is recommended for several reasons. First, Ω1 is tested to ensure that the noise covariance matrices are equal before testing other hypotheses. Otherwise, if Ω1 is false and the noise covariance matrices differ, the sampling distribution of the regression parameters becomes dependent on the unknown population noise covariances. This dependency makes the computation of the sampling distribution challenging, similar to the Behrens–Fisher problem. Next, if differences in noise covariances are not significant, it is recommended to test the equality of AR parameters (Hypothesis Ω2). This simplifies the interpretation of the subsequent Hypothesis Ω3. For instance, the periodic response of Eq. (1) depends on the AR parameters. As a result, differences in the annual cycle response could be due to differences in either the annual cycle parameters or the AR parameters. Testing AR parameters before other parameters resolves this ambiguity, as differences in other parameters are tested only if the differences in AR parameters are not significant.
B8 Diagnosing differences in regression parameters
If is significant, then we conclude that there is a significant difference between the relevant parameters of the VARX models. However, this conclusion does not tell us the nature of those differences. DelSole and Tippett (2022a) proposed diagnosing differences in model parameters based on finding the linear combination of variables that maximizes sub-deviance. This optimization problem leads to an eigenvalue problem whose solutions can be used to compress the sub-deviance into the fewest number of components. In the case of , the interpretation of these components is straightforward: they identify spatial patterns whose ratio of whitened variances is optimized. Further details of this decomposition can be found in DelSole and Tippett (2022a). In contrast, the relationship between the decomposition of and differences in AR parameters, as well as the decomposition of and differences in annual cycle parameters, is not as straightforward. This section aims to clarify the connection between these decompositions and the corresponding parameter differences.
We seek the linear combination of variables that maximizes the sub-deviance for i≥1. Let the coefficients of the linear combination be denoted by , so that the associated variates are
Let us define the variance
and the variance ratio
Because the hypotheses are nested, the residual variances increase with each step in the stepwise procedure, and hence is non-negative. Then the sub-deviance due to regression parameter differences (B23) is
The stationary value of occurs at , which leads to the eigenvalue problem
This eigenvalue problem is identical to Eq. (B21), which shows that eigenvalue problem (B21) is useful both for numerical evaluation and for diagnosing differences in regression parameters. Also, eigenvalue problem (B30) is precisely the eigenvalue problem that arises in covariance discriminant analysis (CDA; DelSole and Tippett, 2022a). The properties of the solutions have been discussed in detail in DelSole and Tippett (2022a). Here we merely summarize the properties that are relevant to diagnosing differences in regression parameters.
Let the eigenvalues of Eq. (B30) be ordered from largest to smallest (), let the corresponding eigenvectors be , and define . The vectors are called loading vectors. Collect the eigenvectors and loading vectors into the matrices
Without loss of generality, the eigenvectors are normalized to satisfy
Since , the normalization implies that the columns of QY and PY form a bi-orthogonal set: . Then, CDA decomposes the covariance matrices into the forms
where S is a diagonal matrix with diagonal elements . Substituting these decompositions into the sub-deviance (B23) and using standard properties of determinants yields
Comparison with Eq. (B29) shows that each term in the sum is a deviance between variates.
The above results show that CDA decomposes sub-deviance into a sum of deviances between variates. The sampling distribution of the leading eigenvalue can be estimated as a byproduct of the Monte Carlo simulations discussed in Sect. B6. However, the interpretation of the decomposition in terms of differences in regression parameters is not apparent. In Appendix C, we show that
where
and is a positive definite matrix defined in Eqs. (C13) and (C17). As a result, vanishes when and is positive otherwise.
Substituting Eq. (B32) into (B30) yields
and are each positive definite and therefore possess symmetric square roots and that are invertible. Thus, the eigenvalue problem can be manipulated into the form
where
Recall that the eigenvalues of are the squared singular values of . Let the SVD of be
where U and V are orthogonal matrices and S is a diagonal matrix with non-negative diagonal elements .
Solving Eq. (B35) for yields
where, following DelSole and Tippett (2022a), we define
These quantities satisfy the identities in Table B2.
A particularly instructive decomposition that follows from Eq. (B36) is
Let pX,j and pY,j denote the jth columns of PX and PY, respectively. Then Eq. (B37) may be interpreted as multiplying the “initial condition” pX,j by the difference in regression parameters to produce a difference in the “one-step response” sjpY,j. Furthermore, the initial condition and response vectors satisfy
(see Table B2). Identities (B38) imply that the initial condition pX,j has a constant probability density from a normal distribution with covariance matrix , and the response pY,j has a constant probability density from a normal distribution with covariance matrix . In this sense, the initial condition and response are “equally likely” realizations from appropriate populations. Then, CDA may be interpreted as finding the initial condition among all equally likely vectors that maximizes the response difference due to the difference in regression parameters.
The above decomposition is sensible when Xi+1 is random, e.g., when the associated coefficients Bi+1 correspond to the autoregressive part of the VARX model. However, if Xi+1 is deterministic, e.g., when it describes the annual cycle, then the predictors are fixed and are usually the same in both data sets (e.g., for annual cycle differences). In this case, a more instructive approach is to propagate regression parameter differences into the time domain as
In terms of decomposition (B36),
where we define
The columns of give the time series by which one should multiply the columns of PY to reconstruct the difference in responses. Thus, decomposition (B40) is similar to principal component analysis. For example, the columns of are time series, and the columns of PY are spatial patterns. Time series for individual models are obtained by computing
so that
The purpose of this Appendix is to prove that and are independent, to derive their distributions, and to show that the latter matrix is a quadratic form of differences in regression parameters for i≥1. The latter result does not seem to have been demonstrated previously. We begin by noting that the noise covariance matrix (B14) can be written equivalently as
where 𝕀 is the identity matrix and ℙi is an orthogonal projection matrix appropriate to Hypothesis Ωi, i.e.,
The projection matrices satisfy (DelSole and Tippett, 2022b)
Consider the telescoping identity
Multiplying this identity by 𝕐T on the left and by 𝕐 on the right and dividing it by gives
where
Because of Eq. (C2), the product of any two terms in parentheses on the right of Eq. (C3) vanishes (i.e., they are “orthogonal”). It follows from Cochran's theorem (Fujikoshi et al., 2010, Sect. 2.4) that are independent and that
We now show that can be written as an explicit function of regression parameter differences. Describing parameter differences through turns out to be awkward because the dimension of depends on Ωi. Accordingly, we define a regression parameter matrix containing all the regression coefficients, and then have the same dimension as but inherit the constraints . For instance, for corresponding to Eq. (B12),
Note that . Estimates of are denoted as , respectively.
In this notation, , and therefore
Furthermore, since ℙi satisfies Eq. (C2),
which shows explicitly that measures the difference in regression parameters. Nevertheless, the interpretation is still obscure because involves a difference of regression parameters under two hypotheses rather than a difference under a single hypothesis. Our goal now is to write in terms of differences in regression parameters under a single hypothesis.
Hypothesis Ωi can be expressed as
where Wi is a suitably chosen matrix with linearly independent rows. For instance, for Ω2, the constraint is and
Also, hypotheses are nested, and therefore
for a suitable matrix . For example, for Ω3, the constraints are and , and therefore
Seber and Lee (2003) show that the least-squares estimate of under constraint (C7) is
where, for i>1,
As a consistency check, we note that WiMi=0, and therefore , as required. We define M1=I. Using this notation, Eq. (C6) may be written as
For i=1, this expression becomes
where
and
For i>1, it proves convenient to define these matrices.
We now show that
From Eq. (C11), Mi+1 is
By a simple matrix product,
Recall the standard matrix inverse identity:
It follows that
and therefore
which is the desired result.
Substituting Eq. (C14) into (C12) yields
where
and
The quadratic form can be simplified further using the fact that
Specifically,
Therefore,
This completes the proof of Eq. (C15), which shows that can be written explicitly as a quadratic form in .
R codes for performing the statistical test described in this paper are available at https://github.com/tdelsole/Comparing-Time-Series-Part-5 (last access: 16 June 2023) and https://doi.org/10.5281/zenodo.7068515 (DelSole, 2022).
The data used in this paper are publicly available from the CMIP5 archive at https://esgf-node.llnl.gov/search/cmip5/ (ESGF, 2023).
Both authors participated in the writing and editing of the manuscript. TD performed the numerical calculations.
The contact author has declared that neither of the authors has any competing interests.
The views expressed herein are those of the authors and do not necessarily reflect the views of these agencies. Some textual passages in this work have been modified for clarity using AI tools.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research was supported by the National Oceanic and Atmospheric Administration (grant no. NA20OAR4310401).
This paper was edited by Likun Zhang and reviewed by three anonymous referees.
Alexander, M. A., Matrosova, L., Penland, C., Scott, J. D., and Chang, P.: Forecasting Pacific SSTs: Linear Inverse Model Predictions of the PDO, J. Climate, 21, 385–402, https://doi.org/10.1175/2007JCLI1849.1, 2008. a, b
Anderson, T. W.: An Introduction to Multivariate Statistical Analysis, Wiley-Interscience, ISBN 978-0-471-36091-9, 1984. a, b
Bach, E., Motesharrei, S., Kalnay, E., and Ruiz-Barradas, A.: Local Atmosphere–Ocean Predictability: Dynamical Origins, Lead Times, and Seasonality, J. Climate, 32, 7507–7519, https://doi.org/10.1175/JCLI-D-18-0817.1, 2019. a
Box, G. E. P., Jenkins, G. M., and Reinsel, G. C.: Time Series Analysis: Forecasting and Control, Wiley-Interscience, 4th edn., ISBN 978-1-118-67502-1, 2008. a, b, c
Brockwell, P. J. and Davis, R. A.: Time Series: Theory and Methods, Springer Verlag, 2nd edn., ISBN 0-387-97482-2, 1991. a, b
Brunner, L. and Sippel, S.: Identifying climate models based on their daily output using machine learning, Environ. Data Sci., 2, e22, https://doi.org/10.1017/eds.2023.23, 2023. a
Chapman, D., Cane, M. A., Henderson, N., Lee, D. E., and Chen, C.: A Vector Autoregressive ENSO Prediction Model, J. Climate, 28, 8511–8520, https://doi.org/10.1175/JCLI-D-15-0306.1, 2015. a
Cordeiro, G. M. and Cribari-Neto, F.: An Introduction to Bartlett Correction and Bias Reduction, Springer, https://doi.org/10.1007/978-3-642-55255-7, 2014. a
DelSole, T.: tdelsole/Comparing-Annual-Cycles: ComparingCyclesv1.0, Zenodo [code], https://doi.org/10.5281/zenodo.7068515 (last access: 30 November 2023), 2022. a
DelSole, T. and Tippett, M.: Statistical Methods for Climate Scientists, Cambridge University Press, Cambridge, https://doi.org/10.1017/9781108659055, 2022a. a, b, c
DelSole, T. and Tippett, M. K.: Laplacian Eigenfunctions for Climate Analysis, J. Climate, 28, 7420–7436, https://doi.org/10.1175/JCLI-D-15-0049.1, 2015. a
DelSole, T. and Tippett, M. K.: Comparing climate time series – Part 1: Univariate test, Adv. Stat. Clim. Meteorol. Oceanogr., 6, 159–175, https://doi.org/10.5194/ascmo-6-159-2020, 2020. a, b
DelSole, T. and Tippett, M. K.: Comparing climate time series – Part 2: A multivariate test, Adv. Stat. Clim. Meteorol. Oceanogr., 7, 73–85, https://doi.org/10.5194/ascmo-7-73-2021, 2021. a
DelSole, T. and Tippett, M. K.: Comparing climate time series – Part 3: Discriminant analysis, Adv. Stat. Clim. Meteorol. Oceanogr., 8, 97–115, https://doi.org/10.5194/ascmo-8-97-2022, 2022a. a, b, c, d, e, f
DelSole, T. and Tippett, M. K.: Comparing climate time series – Part 4: Annual cycles, Adv. Stat. Clim. Meteorol. Oceanogr., 8, 187–203, https://doi.org/10.5194/ascmo-8-187-2022, 2022b. a, b, c, d, e, f, g, h
ESGF: ESGF Meta Grid – CMIP 5, ESGF [data set], https://esgf-node.llnl.gov/search/cmip5/ (last access: 11 September 2019), 2023. a
Eyring, V., Gillett, N., Rao, K. A., Barimalala, R., Parrillo, M. B., Bellouin, N., Cassou, C., Durack, P., Kosaka, Y., McGregor, S., Min, S., Morgenstern, O., and Sun, Y.: Human Influence on the Climate System, in: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, edited by: Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S. L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M. I., Huang, M., Leitzell, K., Lonnoy, E., Matthews, J. B. R., Maycock, T. K., Waterfield, T., Yelekci, O., Yu, R., and Zhou, B., Cambridge University Press, https://doi.org/10.1017/9781009157896.005, 2021. a, b
Fujikoshi, Y., Ulyanov, V. V., and Shimizu, R.: Multivariate Statistics: High-dimensional and Large-Sample Approximations, John Wiley and Sons, ISBN 978-0-470-53987-3, 2010. a, b
Hogg, R. V.: On the Resolution of Statistical Hypotheses, J. Am. Stat. A., 56, 978–989, 1961. a
Hogg, R. V., McKean, J. W., and Craig, A. T.: Introduction to Mathematical Statistics, Pearson Education, 8th edn., ISBN-13 9780137530687, 2019. a
Huang, B., Thorne, P. W., Banzon, V. F., Boyer, T., Chepurin, G., Lawrimore, J. H., Menne, M. J., Smith, T. M., Vose, R. S., and Zhang, H.-M.: Extended Reconstructed Sea Surface Temperature, Version 5 (ERSSTv5): Upgrades, Validations, and Intercomparisons, J. Climate, 30, 8179–8205, https://doi.org/10.1175/JCLI-D-16-0836.1, 2017. a
Labe, Z. M. and Barnes, E. A.: Comparison of Climate Model Large Ensembles With Observations in the Arctic Using Simple Neural Networks, Earth Space Sci., 9, e2022EA002348, https://doi.org/10.1029/2022EA002348, 2022. a
Lund, R., Bassily, H., and Vidakovic, B.: Testing Equality of Stationary Autocovariances, J. Time Ser. Anal., 30, 332–348, 2009. a
Lütkepohl, H.: New introduction to multiple time series analysis, Spring-Verlag, https://doi.org/10.1007/978-3-540-27752-1, 2005. a, b, c, d, e
Mardia, K. V., Kent, J. T., and Bibby, J. M.: Multivariate Analysis, Academic Press, ISBN 978-1-118-73802-3, 1979. a
Mosedale, T. J., Stephenson, D. B., Collins, M., and Mills, T. C.: Granger Causality of Coupled Climate Processes: Ocean Feedback on the North Atlantic Oscillation, J. Climate, 19, 1182–1194, https://doi.org/10.1175/JCLI3653.1, 2006. a
Newman, M.: An Empirical benchmark for decadal forecasts of global surface temperature anomalies, J. Climate, 26, 5260–5269, 2013. a
Penland, C. and Sardeshmukh, P. D.: The optimal growth of tropical sea-surface temperature anomalies, J. Climate, 8, 1999–2024, 1995. a, b
Seber, G. A. F.: A Matrix Handbook for Statisticians, Wiley, https://doi.org/10.1002/9780470226797, 2008. a
Seber, G. A. F.: The Linear Model and Hypothesis: A General Unifying Theory, Springer, https://doi.org/10.1007/978-3-319-21930-1, 2015. a, b
Seber, G. A. F. and Lee, A. J.: Linear Regression Analysis, Wiley-Interscience, https://doi.org/10.1002/9780471722199, 2003. a
Taylor, K. E., Stouffer, R. J., and Meehl, G. A.: An Overview of CMIP5 and the Experimental Design, B. Am. Meteorol. Soc., 93, 485–498, 2012. a
Vimont, D. J.: Analysis of the Atlantic Meridional Mode Using Linear Inverse Modeling: Seasonality and Regional Influences, J. Climate, 25, 1194–1212, https://doi.org/10.1175/JCLI-D-11-00012.1, 2012. a
Whitaker, J. S. and Sardeshmukh, P. D.: A Linear Theory of Extratropical Synoptic Eddy Statistics, J. Atmos. Sci., 55, 237–258, 1998. a
Zanna, L.: Forecast skill and predictability of observed North Atlantic sea surface temperatures, J. Climate, 25, 5047–5056, 2012. a
- Abstract
- Introduction
- Description of the problem and method
- Application to North Atlantic variability
- Conclusions
- Appendix A: Likelihood ratio test
- Appendix B: Multivariate test of the equality of regression models
- Appendix C: Additional properties of sample noise covariance matrices
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Financial support
- Review statement
- References
- Article
(10684 KB) - Full-text XML
- Abstract
- Introduction
- Description of the problem and method
- Application to North Atlantic variability
- Conclusions
- Appendix A: Likelihood ratio test
- Appendix B: Multivariate test of the equality of regression models
- Appendix C: Additional properties of sample noise covariance matrices
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Financial support
- Review statement
- References